Abstract
Spinal and bulbar muscular atrophy (SBMA) is characterized by loss of motoneurons and sensory neurons, accompanied by atrophy of muscle cells. SBMA is due to an androgen receptor containing a polyglutamine tract (ARpolyQ) that misfolds and aggregates, thereby perturbing the protein quality control (PQC) system. Using SBMA AR113Q mice we analyzed proteotoxic stress-induced alterations of HSPB8-mediated PQC machinery promoting clearance of misfolded proteins by autophagy. In muscle of symptomatic AR113Q male mice, we found expression upregulation of Pax-7, myogenin, E2-ubiquitin ligase UBE2Q1 and acetylcholine receptor (AchR), but not of MyoD and of two E3-ligases (MuRF-1 and Cullin3). TGFβ1 and PGC-1α were also robustly upregulated. We also found a dramatic perturbation of the autophagic response, with upregulation of most autophagic markers (Beclin-1, ATG10, p62/SQSTM1, LC3) and of the HSPB8-mediated PQC response. Both HSPB8 and its co-chaperone BAG3 were robustly upregulated together with other specific HSPB8 interactors (HSPB2 and HSPB3). Notably, the BAG3:BAG1 ratio increased in muscle suggesting preferential misfolded proteins routing to autophagy rather than to proteasome. Thus, mutant ARpolyQ induces a potent autophagic response in muscle cells. Alteration in HSPB8-based PQC machinery may represent muscle-specific biomarkers useful to assess SBMA progression in mice and patients in response to pharmacological treatments.
Similar content being viewed by others
Introduction
Spinal and bulbar muscular atrophy (SBMA), or Kennedy’s disease, is an inherited X-linked motoneuron disease (MND) characterized by motoneurons loss in the anterior horns of the spinal cord and in the brain stem, together with atrophy of bulbar, facial and limb muscles1,2,3. Dorsal root ganglia sensory neurons are also affected resulting in sensory function alterations4,5,6,7. Despite being classified as MND, there is evidence that also muscle cells are directly affected in SBMA, suggesting that SBMA might be considered a neuromuscular disease rather than a MND8,9,10,11,12,13,14,15,16,17,18.
SBMA has been linked to the expansion of a CAG triplet repeat sequence located in the exon 1 of the androgen receptor (AR) gene19. The CAG tandem repeat codes for a polyglutamine (polyQ) tract in the N-terminus of the AR protein. In the normal population, the polyQ tract ranges from 9 to 37 Qs (average = 22), while in SBMA patients the polyQ tract is longer than 38 Qs, up to 68 contiguous Qs (ARpolyQ)20,21. Although the function of the polyQ tract is still unknown, it has been proposed that the polyQ tract may serve as a transcriptional regulatory domain22,23,24. PolyQ expansions have been described in eight other totally unrelated proteins involved in neurodegenerative diseases (CAG/polyQ diseases)25, suggesting that the polyQ expansion confers a common gain of neurotoxic function(s) to the mutant proteins. Toxicity has been linked to the acquisition of polyQ-induced aberrant protein conformation (misfolding), which leads to protein aggregation. This process may perturb the protein quality control (PQC) system in cells expressing high levels of ARpolyQ. Bulbar and spinal cord motoneurons, dorsal root ganglia neurons and skeletal muscle cells are all postmitotic cells26,27 and express high levels of AR28, which may make them particularly sensitive to the accumulation of ARpolyQ misfolded species. The nature of the selective cell vulnerability in ARpolyQ and the precise motoneuronal versus muscular contribution to disease is still unclear.
There is emerging evidence supporting the concept that muscle is a key component of disease pathogenesis. Expression of a muscle-specific insulin-like growth factor 1 (IGF-1) isoform selectively in muscle attenuated disease progression in SBMA mice29. More recently, using a neurogenic or myogenic tet-On and Cre-loxP based approach to express ARpolyQ in either motor neurons (NeuroAR) or myocytes (MyoAR) in transgenic mice, Monks and coll. have shown motor function defects in NeuroAR mice, but not in MyoAR mice. Expression of ARpolyQ in muscle of MyoAR mice resulted in a more pronounced reduction in the size of fast glycolytic fibers. The expression of muscle BDNF was reduced in androgen-dependent manner in both models, paralleling changes in motor function, thus supporting the notion that growth factors alteration may play a role in muscle in response to ARpolyQ direct or indirect toxicity18. Nonetheless, the overexpression of human wild type AR (wtAR) in the skeletal muscle of wt mice is sufficient to recapitulate several aspects of SBMA11,13,30,31. Moreover, muscle-specific silencing of the mutant ARpolyQ expression in different SBMA mouse models resulted in prolonged survival, thereby proving evidence for a direct effect of ARpolyQ on muscle atrophy32,33.
Muscle is a direct target of the anabolic androgenic action mediated by the AR34,35. ARpolyQ toxicity strictly depends on androgens, possibly because testosterone triggers the switch of ARpolyQ structure from a “non-toxic” to a “toxic” conformation. Aberrant conformational modifications may generate ARpolyQ aggregates, that are not necessarily toxic per se36. Testosterone binding induces nuclear translocation of misfolded ARpolyQ and nucleus is the cellular compartment where ARpolyQ exerts most of its toxic effects37,38. Interestingly, in adipose mesenchymal cells derived from SBMA patients, inhibition of the proteasome, but not of autophagy, resulted in ARpolyQ accumulation into nuclear aggregates39. However, the ARpolyQ protein reduces long-term protein turnover and blocks the cytoplasmic autophagic flux40,41,42,43,44,45,46. Pharmacological restoration of a normal autophagic flux, obtained by the transcription factor EB (TFEB) activator trehalose47, greatly increased the clearance of mutant misfolded ARpolyQ43,44. Autophagy flux blockage has also been reported in SBMA mice40 and the importance of autophagy in SBMA is sustained by several studies40,48. However, the response to ARpolyQ toxicity in SBMA muscle is complex and rather controversial. Autophagy dysregulation occurs in muscle of AR113Q knock-in SBMA mice already in basal conditions, but autophagy dysregulation is exacerbated in starved mice or in those undergoing to extensive physical exercise; these dysregulation include peculiar transcriptional modulation of autophagic genes induced by the physiological antagonists TFEB and ZKSCAN310. Among the genes regulated by TFEB and increased in SBMA mice in basal conditions is that coding for TFEB itself. Notably, TFEB-target genes, including LC3, Vps11, Vps18 and Lamp1, are also upregulated in muscle samples derived from SBMA patients10. Notably, inhibition of Beclin-1-mediated autophagy activation in AR113Q knock-in SBMA mice diminished skeletal muscle atrophy, extending lifespan and ameliorating their phenotype. Conversely, over-activation of autophagy worsened phenotype9. Thus, restoration of normal flux and normalization of dysregulated autophagy are both essential for ARpolyQ clearance in the skeletal muscle of SBMA mice.
Pathways that re-route ARpolyQ to degradation mediated by ubiquitin-proteasome system (UPS) may also be of relevance for its clearance. A delicate equilibrium exists between UPS and autophagy, as both pathways are essential for proper functions of the PQC system42,43,46,49. This equilibrium is regulated by a number of proteins, including the co-chaperones BAG1 (for UPS clearance) and BAG3 (for autophagic clearance)50. In addition, flux restoration and autophagy modulation can be regulated by few specific chaperones, like some members of the small HSP family (HSPBs), such as HSPB8. Notably, mutations in HSPB8 (at K141 with E or N) have been linked to Charcot-Marie-Tooth type 2L disease and hereditary distal motor neuropathy type II (dHMNII)51,52. By interacting with BAG3 (in a 2:1 ratio) HSPB8 decreases ARpolyQ aggregation, by increasing its solubility and clearance43. HSPB8 also acts on several other misfolded proteins involved in neurodegenerative diseases43,49,53,54. HSPB8 acts as facilitator, rather than activator of autophagy43. This requires the interaction of 2x(HSPB8)/BAG3 with HSC70/CHIP, allowing misfolded substrate ubiquitination for its p62 mediated recognition and insertion into autophagosomes53,55. This effect is not associated with mutant dHMNII-HSPB856. In ALS mice, both anterior horn spinal cord motor neurons and the skeletal muscle cells respond to proteotoxicity by activating a robust HSPB8-mediated PQC system response53,57,58. Nothing is known about the involvement of the HSPB8 and BAGs machinery in SBMA skeletal muscle and the identification of specific autophagy related molecular markers of skeletal muscle degeneration in SBMA might represent a diagnostic valuable tool for monitoring the disease progression. For these reasons, in this study, we have provided an extensive characterization of the autophagic activation as well as of the HSPB8 machinery in the PQC system response in SBMA muscle.
Results
Here, we investigated the specific response of the HSPB8-mediated PQC machinery to ARpolyQ toxicity in the skeletal muscle of a knock-in mouse model of SBMA. SBMA mice have been obtained by knocking in a segment of the human exon 1 carrying 113 CAG repeats, into the homolog region of the endogenous mouse AR gene (AR113Q mice)59. Thus, being controlled by the endogenous AR promoter, these mice retained the physiological levels of AR proteins and AR cell localization in all tissues59. For the purpose of our study, this aspect provides a significant advantage over other SBMA mouse models developed using exogenous promoters to drive the unspecific overexpression of exogenous human AR. Over-expression and incorrect cell production of the toxic ARpolyQ might induce unspecific abnormalities in the PQC system response, especially in skeletal muscle tissue, which is particularly sensitive to the presence of altered proteins58,60.
Muscle atrophy and altered motor behavior are overt at 24 weeks of age in AR113Q knock-in mice
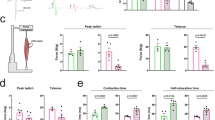
To evaluate disease manifestations in AR113Q knock-in mice, we analyzed spinal cord and muscle pathology at 24 wks of age. In agreement with previous reports9,10,32,59, the histopathological analysis (Fig. 1(a), upper and middle insets) of the anterior horn of the spinal cord excluded the presence of motoneuronal loss in AR113Q male mice at 24 wks of age. This was confirmed by the statistical evaluation of the total number of motoneurons presents at this age (Fig. 1(b)) both in the tissue and in average number for the slice considered (left and right panels, respectively). Conversely, histological analysis of quadriceps muscle (Fig. 1(a), lower insets) revealed signs of myopathy. Hematoxylin and Eosin analysis showed that the quadriceps of AR113Q mice was characterized by the presence of smaller fibers compared to those present in age-matched male control mice. It has been previously showed that the functional deficit on behavioral testing of AR113Q males is also accompanied by morphologic changes in skeletal muscle which are indicative of both neurogenic and myopathic effects in a way that resemble the mixed features described in biopsies of Kennedy’s disease muscle (29, 30). It has been shown that in these mice myopathy occurs before overt spinal cord pathology59,61. In addition, the body weight of AR113Q mice was decreased compared to control mice (9 wks = controls 22.86 ± 0.4417, AR113Q male mice 24.21 ± 0.342; 24 wks = controls 28.45 ± 0.7571, AR113Q male mice 26.75 ± 0.4581) since SBMA mice tended to grow less than control (Fig. 1(c)). To assess motor behavior in comparison with literature data9,10,32,59,61, we performed grip strength (Fig. 1(d)) and rotarod (Fig. 1(e)) analyses. We confirmed that AR113Q male mice do not show motor function abnormalities at 9 wks of age. On the other hand, AR113Q mice showed reduced performance at 24 wks of age (mean ± SEM grip strength test: 9 wks = controls 111.7 ± 18.82, AR113Q male mice 105.4 ± 6.566; 24 wks = controls 101.5 ± 10.53, AR113Q male mice 61.42 ± 3.667 p < 0.001 vs control 24wks; p < 0.001 vs AR113Q 9wks. Rotarod test: 24 wks = controls 430,4 ± 45.14, AR113Q male mice 196.9 ± 36.78, p < 0.05 vs controls). Since changes in the muscle mass may affect the behavioural test results, both the grip strength and the rotarod tests were also statistically evaluated using body weight as covariate; however, the significant variations were similar to those presented above (not showed) indicating that the body weight had no influence on the behavioural assays performed. Interestingly, in AR113Q male mice insoluble forms of mutant ARpolyQ were only detectable in the quadriceps muscle and not in the spinal cord (Fig. 1(f)).
Preliminary characterization of the SBMA mouse model.
(a) Spinal cord and quadricep cross sections of control and AR113Q male mice at 24 weeks of age (corresponding to symptomatic stage). Nissl staining of lumbar spinal cord shows the cell body of lower motor neurons. Hematoxylin-Eosin staining of quadriceps underscores a marked decrease in fiber size of AR113Q mice compared to control mice. (b) Left Panel: average number of total motor neurons in the lumbar spinal cord of control and AR113Q mice at 24 weeks of age (control mice, n = 5; AR113Q, n = 6); Right panel: average number of motor neurons for each slice (number of slices = 7). (c) Measurement of body weight in control and AR113Q mice at 9 and 24 weeks of age (control mice, n = 5; AR113Q, n = 20). (d) Grip strength test in control and AR113Q mice at 9 and 24 weeks of age (control mice, n = 5; AR113Q, n = 15). **p < 0.001 vs. 24 weeks control mice; °°p < 0.001 vs. 9 weeks AR113Q mice. (e) Rotarod test in control and AR113Q male mice at 24 weeks of age. *p < 0.05 vs. 24 weeks control mice (control mice, n = 5; AR113Q, n = 15). (f) Filter retardation assay in spinal cord and quadriceps of control and AR113Q male mice at 24 weeks. Androgen receptor AR aggregation was visible only in ARQ113 quadriceps at 24 weeks of age. **p < 0.001 vs. 24 weeks quadriceps of control mice.
These results indicate that AR113Q mice develop progressive motor dysfunction and have reduced muscle strength.
Disease-associated alteration of muscular markers in symptomatic AR113Q male mice
In order to elucidate which changes in muscle dynamics occur in pre-symptomatic (9 wks) and symptomatic (24 wks) AR113Q mice, we analyzed the expression of proliferation/differentiation and/or atrophy markers in these animals. As an additional control, we also compared the results obtained in symptomatic AR113Q male mice with those obtained in age-matched AR113Q females. Females do not develop the pathology because they have lower levels of androgens in the serum compared to male mice10,32,59,61. The results of this first set of analyses are shown in Fig. 2. No changes in MyoD expression were observed in muscle at all ages considered (Fig. 2(a)). Conversely, MyoG (myogenin) expression was significantly increased up to three-fold in symptomatic (24 wks) AR113Q male mice over controls (e.g.: Wt mice at all ages, AR113Q male mice at presymptomatic stage 9 wks and AR113Q female mice at 24 wks) (Fig. 2(b). We next analysed the expression of Pax-7 (a regulator of myogenesis acting on the proliferation of muscle precursors) and we found that the expression of this gene tend to decrease with age in control mice but in AR113Q male mice at 24 wks the Pax-7 mRNA were comparable to that of younger control mice (Fig. 2(c)) thus suggesting that in symptomatic animals muscle responds with an attempt to maintain elevated the myogenic pathway.
Expression of muscular markers in quadricep muscles of SBMA mouse model.
RT-qPCRs were performed on total RNA extracted from quadricep muscles of male non-transgenic (Wt) mice and of AR113Q mice at 9 (corresponding to presymptomatic stage) or 24 (corresponding to symptomatic stage) weeks of age. As additional control, quadriceps of female AR113Q mice at 24 weeks of age were used (n = 5). All animals were age-matched. Data have been normalized to the amount of GAPDH mRNA, expressed relative to the levels determined in Wt mice at 9 weeks taken as internal reference and expressed as fold changes. Data are means ± SD of three independent replicates for Wt and AR113Q mice at 9 weeks and of five independent replicates for Wt and AR113Q mice at 24 weeks. (A) RT-qPCR on MyoD mRNA expression levels in quadricep muscles. (B) RT-qPCR on MyoG mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice; °p < 0.05 vs. 9 weeks AR113Q mice. (C) RT-qPCR on Pax7 mRNA expression levels in quadricep muscles. §p < 0.05 vs. 9 weeks Wt mice; *p < 0.05 vs. 24 weeks Wt mice. (D) RT-qPCR on Atrogin-1 mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice. (E) RT-qPCR on MURF-1 mRNA expression levels in quadricep muscles. (F) RT-qPCR on Cullin3 mRNA expression levels in quadricep muscles. (G) RT-qPCR on UBE-2Q mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice; °°p < 0.001 vs. 9 weeks AR113Q mice. (H) RT-qPCR on TGFβ1 mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice; °p < 0.05 vs. 9 weeks AR113Q mice. (I) RT-qPCR on PGC-1α mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice; °°p < 0.001 vs. 9 weeks AR113Q mice. (L) RT-qPCR on AchR mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice; °p < 0.05 vs. 9 weeks AR113Q mice.
Given that AR113Q male mice show signs of muscle atrophy at 24 wks of age and are considered as symptomatic, we analysed whether this correlates with increased protein turnover mediated by the ubiquitin proteasome system (UPS). In particular, we analysed atrogin-1 and muscle RING-finger protein-1 (MuRF-1), which are two E3 ubiquitin ligases, with important regulatory functions on ubiquitin-mediated protein degradation in skeletal muscle. In addition, these two E3 ubiquitin ligases play a central role in controlling muscle size62. In the case of atrogin-1 expression, which in muscle it is known to increase with age63, we found an unexpected significant down-regulation in symptomatic (24 wks) AR113Q male mice, compared to age-matched controls (Wt male mice at 24 wks) (Fig. 2(d)). These data suggest defects in the activation of the UPS in AR113Q mice, even though MuRF-1 expression remained unchanged (Fig. 2(e)). According to this gene expression analysis UPS seems to be not massively up-regulated in these mice, as already postulated in previous reports10. We also analysed the expression of a third E3 ubiquitin ligase, the cullin-RING ubiquitin ligase (Cullin3), whose function is relevant for cellular stress response in a number of diseases, including neurodegenerative diseases64 and dominant distal myopathy65. However, no changes of Cullin3 expression were detected in AR113Q mice (Fig. 2(f)). Therefore, despite the fact that at 24 wks of age AR113Q male mice already show symptoms of the disease accompanied by signs of muscle atrophy and MyoG activation, the catabolic activity of muscle proteins mediated by atrogin-1, MuRF-1 and Cullin-3 ubiquitin ligases did not take place yet. On the other hand, in symptomatic (24 wks) AR113Q male mice, we surprisingly found that the expression of E2-ubiquitin ligase, the Ubiquitin-Conjugating Enzyme E2Q Family Member 1 (UBE2Q1) (Fig. 2(g)), was up-regulate.
Next, we analysed the expression of TGFβ1, a marker for muscle fiber damage or atrophy. We found that the expression of TGFβ1 is specifically upregulated only in the symptomatic (24 wks) AR113Q male mice (Fig. 2(h)), paralleling the expected deleterious effects of mutant ARpolyQ on muscle fibers of these mice. This is in line with the observation that, in a tet-repressible muscle-specific TGFβ1 transgenic mouse, the induction of TGFβ1 overexpression resulted in muscle weakness and atrophy, with the appearance of endomysial fibrosis and smaller myofibers66, suggesting the existence of a primary effect of TGFβ1 on muscle atrophy.
In addition, we observed a robust increase of PGC-1α mRNA expression specifically in the symptomatic (24 wks) AR113Q male mice (Fig. 2(i)). PGC-1α is a transcriptional co-activator (i.e. of PPARγ) and regulates energy metabolism and mitochondrial biogenesis and functions. PGC-1α has been already implicated in muscle dysfunction in amyotrophic lateral sclerosis (ALS)67,68. Finally, by analysing the level of the denervation marker AchR, we found a dramatic increase of AchR gene transcription in the symptomatic (24 wks) AR113Q male mice compared to all controls mice tested as comparison (Fig. 2(l)). AchR and MyoG genes are both induced after denervation32. Notably, their up-regulation occurs in the absence of motoneuron loss (See Fig. 1). Collectively, these data show that, in AR113Q mice markers of muscle damage are upregulated in symptomatic animals.
Correlation between symptoms and AR expression in muscle and in spinal cord of AR113Q male mice
Next, we asked whether ARpolyQ induces, during disease progression, a negative selection of muscle cells poorly expressing the toxic protein as a compensatory mechanism linked to detrimental effects of ARpolyQ (Fig. 1). To test our hypothesis, we analysed the levels of AR mRNA transcript and protein in skeletal muscle. The data on the analysis of AR expression are shown in Fig. 3(a). As it clearly appears in the graphs, AR expression is equivalent in all samples considered, including those derived from AR113Q male mice at 24 wks. The lack of variation in AR expression in symptomatic AR113Q male mice indicates that apparently no negative selective pressure is induced by ARpolyQ toxicity in muscle cells.
Expression of AR in quadriceps muscle and spinal cords of SBMA mouse model.
RT-qPCRs were performed on total RNA extracted from quadricep muscles (A) or whole spinal cords (B) of male non-transgenic (Wt) mice and of AR113Q mice at 9 (corresponding to presymptomatic stage) or 24 (corresponding to symptomatic stage) weeks of age. As additional control, quadriceps muscles of female AR113Q mice at 24 weeks were used (n = 5). Data have been normalized to the amount of GAPDH mRNA, expressed relative to the levels determined in Wt mice at 9 weeks taken as internal reference and expressed as fold changes. Data are means ± SD of three independent replicates for Wt and AR113Q mice at 9 weeks and of five independent replicates for Wt and AR113Q mice at 24 weeks. (A) RT-qPCR on AR mRNA expression levels in quadricep muscles. (B) RT-qPCR on AR mRNA expression levels in spinal cords.
From the anatomopathological and morphological analysis reported in Fig. 1, no motoneuronal loss is present in the anterior horn of the spinal cord of AR113Q male mice at 24 wks of age. However, since symptom manifestations could also be due to its neurotoxicity at motoneuronal levels, we also decided to analyse the expression of ARpolyQ in the spinal cord of the same mice, to include molecular evidence that AR expression remains unchanged in this structure. In fact, AR expression (together with the androgen-activator machinery) in the spinal cord is mainly confined to the anterior horn motoneurons28,69,70 (affected in SBMA patients), while interneurons and glial cells express very low levels of AR71,72,73. Thus, it is conceivable that AR mRNA variations in spinal cord correlate with the motoneuronal cell modification and are representative of motoneuron loss. The data reported in Fig. 3(b) show a mild, but not significant decrease in AR expression in AR113Q male mice at 24 wks. From these data we conclude that expression of AR in AR113Q is similar to that of control mice both in muscle and spinal cord.
Modification of autophagic markers in muscle of symptomatic AR113Q male mice
To better understand whether symptom onset correlates with an activation of gene expression of proteins involved in PQC system in muscle cells, we measured changes in the expression of autophagic markers in the skeletal muscle of symptomatic AR113Q male mice. It has already been reported that the autophagy master regulator gene product TFEB is up-regulated in these mice when symptoms appear10. TFEB increase is paralleled by the increase of some specific TFEB target genes10. Here, we analysed the expression of four genes encoding proteins which are essential for autophagy initiation and are critical markers for the autophagic process. Two of these genes are critical for autophagosome assembly: Beclin-1 (the ATG6 ortholog, which regulates and associates to Vps34 to induce autophagy) and ATG10 (an E2-like enzyme involved in LC3 activation and modification essential for autophagosome formation); the other two, are TFEB regulated and considered typical autophagy markers: p62/SQSTM1 (which recognizes ubiquitinated protein for their insertion into autophagosomes) and LC3 (which after its lipidation to LC3-II associates to the autophagosome membrane)43,44. We found that all autophagic markers analysed were robustly and specifically induced in skeletal muscle of symptomatic AR113Q male mice (Beclin-1 in Fig. 4(a); ATG10 in Fig. 4(b); p62/SQSTM1 in Fig. 4(c); LC3 in Fig. 4(d)) over controls; no changes were present by comparing muscles of all other mice groups included in the study.
Expression of autophagic markers in quadriceps muscle of SBMA mouse model.
RT-qPCRs were performed on total RNA extracted from quadricep muscles of male non-transgenic (Wt) mice and of AR113Q mice at 9 (corresponding to presymptomatic stage) or 24 (corresponding to symptomatic stage) weeks of age. As additional control, quadriceps of female AR113Q mice at 24 weeks were used (n = 5). All animals were age-matched. Data have been normalized to the amount of GAPDH mRNA, expressed relative to the levels determined in Wt mice at 9 weeks taken as internal reference and expressed as fold changes. Data are means ± SD of three independent replicates for Wt and AR113Q mice at 9 weeks and of five independent replicates for Wt and AR113Q mice at 24 weeks. (A) RT-qPCR on Beclin1 mRNA expression levels in quadricep muscles. *p < 0.05 vs. 24 weeks Wt mice; °°p < 0.001 vs. 9 weeks AR113Q mice. (B) RT-qPCR on ATG10 mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice; °°p < 0.001 vs. 9 weeks AR113Q mice. (C) RT-qPCR on P62/SQSTM1 mRNA expression levels in quadricep muscles . **p < 0.001 vs. 24 weeks Wt mice; °p < 0.05 vs. 9 weeks AR113Q mice. (D) RT-qPCR on LC3 mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice; °p < 0.05 vs. 9 weeks AR113Q mice.
Induction of pro-autophagic chaperones in muscle of symptomatic AR113Q male mice
We already demonstrated that one of the key protein involved in the autophagic removal of misfolded proteins (such as ARpolyQ associated to SBMA or mutant SOD1 and TDP-43 associated to ALS, etc) is the HSPB8. Expression of HSPB8 is robustly increased by UPS blockage to facilitate autophagy53,54. HSPB8 expression is also enhanced when PQC is altered in ALS especially in motoneuron and muscle cells57,58. Therefore, we investigated whether, also in the case of SBMA, HSPB8 may be involved in the response to protein toxicity in muscle. When we analysed HSPB8 expression in SBMA mice (Fig. 5(a)) we found that this gene was transcriptionally up-regulated specifically in symptomatic AR113Q male mice, while it remained unchanged in all other mice tested and at all time points considered. HSPB8 interacts with another member of its same family, namely the chaperone HSPB2. Interestingly, HSPB2 usually conjugates with a third member of the family, the chaperone HSPB3, found mutated in an axonal type of motor neuropathy74; HSPB2 and HSPB3 are capable to form hetero-oligomers in a 3:1 subunit ratio75 and are both highly expressed in skeletal muscle where they play relevant roles during myogenic differentiation76,77. Thus, we analysed whether the induction of HSPB8 is paralleled by the induction of these other two HSPBs in response to ARpolyQ muscle toxicity. Interestingly, both genes were significantly upregulated in the muscle of symptomatic AR113Q male mice, respect all the other mice considered (Fig. 5(b,c)). Notably, these chaperones were selectively upregulated in male, but not female AR113Q mice, indicating that their upregulation mirrors androgen-dependent effects of ARpolyQ expression.
Expression of pro-autophagic chaperones in quadriceps muscle of SBMA mouse model.
RT-qPCRs were performed on total RNA extracted from quadricep muscles of male non-transgenic (Wt) mice and of AR113Q mice at 9 (corresponding to presymptomatic stage) or 24 (corresponding to symptomatic stage) weeks of age. As additional control, quadriceps of female AR113Q mice at 24 weeks were used (n = 5). All animals were age-matched. Data have been normalized to the amount of GAPDH mRNA, expressed relative to the levels determined in Wt mice at 9 weeks taken as internal reference and expressed as fold changes. Data are means ± SD of three independent replicates for Wt and AR113Q mice at 9 weeks and of five independent replicates for Wt and AR113Q mice at 24 weeks. (A) RT-qPCR on HSPB8 mRNA expression levels in quadricep muscles. *p < 0.05 vs. 24 weeks Wt mice; °°p < 0.001 vs. 9 weeks AR113Q mice. (B) RT-qPCR on HSPB2 mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice; °°p < 0.001 vs. 9 weeks AR113Q mice. (C) RT-qPCR on HSPB3 mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice; °°p < 0.001 vs. 9 weeks AR113Q mice.
To exert its action, HSPB8 must associate to its co-chaperone BAG3. Once misfolded species are recognized by HSPB8-BAG3, they interact with HSC70-CHIP and the misfolded substrates become ubiquitinated for p62 recognition and autophagosome insertion49,53,55. HSPB8 is highly unstable in the absence of BAG3. In addition, when low level of BAG3 are present and the other co-chaperon BAG1 is induced, BAG1 preferentially associates to HSC70-CHIP to re-route protein degradation via the UPS, rather than autophagy42,49,78,79. The ratio BAG3:BAG1 is thus essential to select which PQC degradative pathway is preferred to clear misfolded proteins in a given cell type. BAG1 and BAG3 expression was then analysed in muscles of Wt mice and SBMA mice at all ages considered. We found that both co-chaperones were induced in the presence of mutant ARpolyQ in muscle of symptomatic (24 wks) AR113Q male mice, but not in all other controls tested (Fig. 6(a): BAG1 in, Fig. 6(b): BAG3). However the ratio measured was dramatically increased in favor of BAG3 (Fig. 6(c): BAG3:BAG1 ratio higher than 3) in symptomatic AR113Q male mice, while it remained similar to Wt mice in all other controls tested. Therefore, in symptomatic muscles, the autophagic pathway is preferentially activated in response to mutant ARpolyQ protein toxicity.
Expression of BAG1 and BAG3 co-chaperones in quadriceps muscle of SBMA mouse model.
RT-qPCRs were performed on total RNA extracted from quadricep muscles of male non-transgenic (Wt) mice and of AR113Q mice at 9 (corresponding to presymptomatic stage) or 24 (corresponding to symptomatic stage) weeks of age. As additional control, quadriceps of female AR113Q mice at 24 weeks were used (n = 5). All animals were age-matched. Data have been normalized to the amount of GAPDH mRNA, expressed relative to the levels determined in Wt mice at 9 weeks taken as internal reference and expressed as fold changes. Data are means ± SD of three independent replicates for Wt and AR113Q mice at 9 weeks and of five independent replicates for Wt and AR113Q mice at 24 weeks. (A) RT-qPCR on BAG1 mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice; °p < 0.05 vs. 9 weeks AR113Q mice. (B) RT-qPCR on BAG3 mRNA expression levels in quadricep muscles. **p < 0.001 vs. 24 weeks Wt mice; °°p < 0.001 vs. 9 weeks AR113Q mice. (C) BAG3:BAG1 relative ratio of mRNA expression levels in quadricep muscles. Data represent variations of the relative levels of BAG3 and BAG1 normalized over the relative BAG3 and BAG1 levels of age-matched 9 weeks Wt mice. **p < 0.001 vs. 24 weeks Wt mice; °°p < 0.001 vs. 9 weeks AR113Q mice.
Modification of proteins regulating autophagy and HSPB8-mediated response in skeletal muscle of symptomatic AR113Q male mice
We next evaluated whether the observed changes in gene expression of transcripts involved in regulating autophagy and HSPB8-mediated response are also paralleled by variations at protein levels. Western blot analyses (Fig. 7(a)) showed that despite the increased expression (see Fig. 4(c)), the overall protein levels of p62/SQSTM1 remained unchanged in symptomatic AR113Q male mice as compared to control wt mice of the same age (quantification in Fig. 7(b)). This is not surprising since once autophagy is activated by the expression of TFEB regulated genes (including p62/SQSTM1), the p62/SQSTM1 protein recognizes ubiquitinated substrates for their engulfment into autophagosomes and p62 itself is processed by autophagy. Thus, p62/SQSTM1 steady state levels depend upon its transcriptional induction (during autophagy activation) and its turnover (during autophagy progression)43,44,53. The fact that p62/SQSTM1 does not accumulate into p62/SQSTM1 bodies suggests that the overall autophagic flux is not blocked at this initial stage by the presence of the mutant ARpolyQ in skeletal muscle. This finds further support in the observation that also LC3 expression (see Fig. 4(d), along with Beclin-1 (Fig. 4(a) and ATG10 (Fig. 4(b)) is robustly induced, proving that autophagy is activated in skeletal muscle of symptomatic AR113Q male mice. However, the overall levels of the total LC3-I are greatly reduced (Fig. 7(c)) while the LC3-II/LC3-I ratio (Fig. 7(d)) are significantly increased in skeletal muscle of symptomatic AR113Q male mice. Notably, also LC3 (after its lipidation to LC3-II) is inserted into autophagosomes to be cleared (with p62/SQSTM1 and the ubiquitinated substrated to be degraded) when autophagic flux is sustained into cells43,44,53. Thus, these data clearly indicate not only that autophagy is highly activated (as evidenced by increased expression of the autophagy markers reported in Fig. 4), but it is not stalled (as evidenced by reduced p62/SQSTM1 and LC3 protein levels and increased LC3-II/LC3-I ratio) in skeletal muscle of symptomatic AR113Q male mice. The proper activity of the autophagic flux may be maintained by the increased expression of the entire HSPB8-related machinery (which includes HSPB8 itself (Fig. 5(a)), BAG3 (Fig. 6(b)) and possibly the other small HSPs, HSPB2 (Fig. 5(b)) and HSPB3 (Fig. 5(c)) and also by the HSPB8 protein levels (Fig. 7(e)) in skeletal muscle of symptomatic AR113Q male mice as compared to wt male mice. Increased HSPB8 levels are accompanied by the increased levels of its co-chaperone BAG3 (Fig. 7(f)), which is absolutely required to stabilize HSPB8 protein43,49,53,54 and the increased BAG3:BAG1 ratio (Fig. 7(g,h)) which is indicative of the misfolded substrate re-routing from the UPS to autophagy53,57,58.
Evaluation of the levels of proteins regulating autophagy and HSPB8-mediated response in skeletal muscle of symptomatic AR113Q male mice.
(A) Western blot analysis perfomed on quadriceps of male wt and AR113Q mice at 9 and 24 weeks. As additional control, quadriceps of female AR113Q mice at 24 weeks were used. (B) Quantification of p62/SQSTM1 protein expression levels in in wt (n = 3) and AR113Q (n = 3) male mice at 24 weeks. The p62/SQSTM1 protein levels were normalized using tubulin as control. (C) Quantification of LC3-I protein expression levels in in wt (n = 3) and AR113Q (n = 3) male mice at 24 weeks. The LC3-I protein levels were normalized using tubulin as control. *p < 0.05 vs. 24 weeks Wt mice. (D) Quantification of the LC3-II/LC3-I ratio in wt (n = 3) and AR113Q (n = 3) male mice at 24 weeks. (E) Quantification of HSPB8 protein expression levels in in wt (n = 3) and AR113Q (n = 3) male mice at 24 weeks. The HSPB8 protein levels were normalized using tubulin as control; *p < 0.05 vs. 24 weeks Wt mice. (F) Quantification of BAG3 protein expression levels in in wt (n = 3) and AR113Q (n = 3) male mice at 24 weeks. The BAG3 protein levels were normalized using tubulin as control; *p < 0.05 vs. 24 weeks Wt mice. (G) Quantification of BAG1 protein expression levels in in wt (n = 3) and AR113Q (n = 3) male mice at 24 weeks. The BAG1 protein levels were normalized using tubulin as control. (H) Quantification of the ratio between BAG3 and BAG1 protein levels in wt (n = 3) and AR113Q (n = 3) male mice at 24 weeks; **p < 0.001 vs. 24 weeks Wt mice.
The autophagic response in spinal cord of AR113Q male mice
We then evaluated whether changes in PQC markers are also present in the spinal cords of the AR113Q mice at the pre-symptomatic and symptomatic stages. When we analysed the expression of Beclin-1, p62/SQSTM1, LC3, HSPB8, HSPB2, HSPB3, BAG1 and BAG3 (Fig. 8(a–h), respectively) we failed to detect variation in the different groups tested, including symptomatic (24 wks) AR113Q male mice. The only variation observed was found in the relative BAG3:BAG1 ratio (Fig. 8(i)), which in symptomatic AR113Q male mice was in favor of the BAG3 expression suggesting the existence of a preferential activation of the autophagic pathway over the UPS pathway also in the spinal cord.
Expression of autophagic markers, chaperones and co-chaperones in spinal cords of SBMA mouse model.
RT-qPCRs were performed on total RNA extracted from whole spinal cord of male non-transgenic (Wt) mice and of AR113Q mice at 9 weeks (corresponding to presymptomatic stage) or 24 weeks (corresponding to symptomatic stage). As additional control, spinal cord of female AR113Q mice at 24 weeks were used (n = 5). All animals were age-matched. Data have been normalized to the amount of GAPDH mRNA, expressed relative to the levels determined in Wt mice at 9 weeks taken as internal reference and expressed as fold changes. Data are means ± SD of three independent replicates for Wt and AR113Q mice at 9 weeks and of five independent replicates for Wt and AR113Q mice at 24 weeks. (A) RT-qPCR on Beclin1 mRNA expression levels in spinal cord. (B) RT-qPCR on P62/SQSTM1 mRNA expression levels in spinal cord. (C) RT-qPCR on LC3 mRNA expression levels in spinal cord. (D) RT-qPCR on HSPB8 mRNA expression levels in spinal cord. (E) RT-qPCR on HSPB2 mRNA expression levels in spinal cord. (F) RT-qPCR on HSPB3 mRNA expression levels in spinal cord. (G) RT-qPCR on BAG1 mRNA expression levels in spinal cord. (H) RT-qPCR on BAG3 mRNA expression levels in spinal cord. (I) BAG3:BAG1 relative ratio of mRNA expression levels in whole spinal cord. Data represent variations of the relative levels of BAG3 and BAG1 normalized over the relative BAG3 and BAG1 levels of age-matched 9 weeks Wt mice. *p < 0.05 vs. 24 weeks Wt mice.
Discussion
In the present study, we have analysed the role of the PQC system response, mediated by the HSPB8 machinery and alternative pathways in the skeletal muscle and spinal cord of mice expressing physiological levels of mutant ARpolyQ. All the gene expression analyses on the symptomatic AR113Q male mice have been performed in comparison to multiple controls, which included normal male mice and pre-symptomatic AR113Q male mice, as well as age-matched non-symptomatic AR113Q female mice, which are non-symptomatic.
Using histological analysis and behavioral test, we demonstrated that AR113Q show symptoms by 24 wks of age. At this age the AR113Q male mice showed signs of muscle atrophy, but no motoneuronal loss in the anterior horn spinal cord was detected, as demonstrated by the histopathological analysis performed on spinal cord sections. These data are in agreement with previous reports10,59,61.
We analyzed several markers of muscle atrophy. We found that MyoG (or myogenin) expression was greatly up-regulated in symptomatic AR113Q male mice, while the expression of Pax-7 was found to be decreased over time in wt mice, but was maintained at elevated levels also at older age in symptomatic AR113Q mice. Interestingly, elevated Pax-7 expression is indicative of an attempt to regenerate after muscle injury. Together with the observation of elevated levels of AchR, these data confirmed alteration at muscular levels and possibly an association to denervation (even if not accompained by motoneuronal loss) in the skeletal muscle of AR113Q mice9,30,32,80.
Instead, no changes were observed in the expression of the differentiation marker MyoD (as expected80) or the E3 ubiquitin ligases MuRF-1 and Cullin3 in the muscle of AR113Q and control mice at all ages tested. Unexpectedly, Atrogin-1 was found significantly down-regulated in symptomatic (24 wks) AR113Q male mice, possibly in response to an already activated atrophic process. It remains to be elucidated whether this atrophic process is mediated by unknown pathways involving the E2-ubiquitin ligase UBE2Q1, which is dramatically up-regulated in the muscle of symptomatic AR113Q male mice.
We then wondered whether symptomatic AR113Q male mice were characterized by modification of selected markers that we found altered in tg ALS mouse models, such as TGFβ1 (which correlates with muscle fiber damage or atrophy)34,81, or PGC-1α67 (which regulates energy metabolism and mitochondria biogenesis and functions). Notably, both genes were dramatically up-regulated specifically in affected mice. This finding suggests a primary role of TGF-β1 on muscle atrophy. In fact, when TGF-β1 is overexpressed it induces endomysial fibrosis and promotes myofibers atrophy66 accompanied by the ability of muscle cell to maintain a correct metabolism under these conditions and an attempt to overload the system by enhancing the PGC-1α/PPARγ pathway. A role for TGF-β1 in SBMA pathology has also been proposed by other Authors demonstrating that, in the spinal cord of male transgenic mice carrying the mutant human AR, a reduced expression of the type II receptor for TGF-β and a hampered nuclear translocation of pSmad2/3 proteins are present82. The blockage of the TGF-β signaling could be responsible for the motoneuron damage in SBMA since this growth factor is known to have a crucial role in the survival and function of adult neurons83. Despite these relevant variations in muscle in response to mutant ARpolyQ in symptomatic AR113Q male mice, we did not observe changes in the expression levels of the AR mRNA, strongly suggesting the absence of either positive (linked to trophic androgens/AR effects) or negative (linked to cytotoxic androgens/AR effects) selection of muscle cell at this relatively early stages of disease.
We then analysed whether in our colony of AR113Q mice alterations of the autophagic pathways were detectable at pre-symptomatic or symptomatic stages in male mice. It has already been reported that the autophagy master regulator TFEB is upregulated in muscles of SBMA mice and patients10. Here we found a mild, but not significant, up-regulation of TFEB in the symptomatic AR113Q male mice as compared to the control used (data not shown). Despite this, we found that the gene expression of all other autophagic markers analysed (Beclin-1, ATG10, p62/SQSTM1, LC3) was dramatically increased in the muscle of symptomatic AR113Q male mice. Interestingly, with regards to p62/SQSTM1 and LC3 autophagic markers, Chua and coll10 reported that the insoluble (but not the soluble) fraction of the p62/SQSTM1 protein and of the LC3 autophagosome-anchored lipidated form (LC3-II) were greatly enhanced in their symptomatic AR113Q male mice. In our model, we also found not only an increased p62/SQSTM1 and LC3 mRNAs expression, but a stabilization (for p62/SQSTM1) or a reduction (for LC3) in the overall levels of the corresponding coded proteins. Interestingly, both p62/SQSTM1 and LC3 proteins are normally processed by autophagy when autophagic flux is properly activated and it is not blocked. Notably, the increased turnover of total LC3 protein was accompanied by an increased LC3-II/LC3-I ratio. Thus, in agreement with a previous report10, collectively, these data are strongly suggestive of autophagy activation sustained by a regular flux in atrophic skeletal muscle of symptomatic AR113Q male mice.
On these bases, we demonstrated that the HSPB8-mediated PQC machinery is also greatly activated in the muscle of symptomatic AR113Q male mice. In fact, the levels of both the mRNA and protein levels of HSPB8 and its co-chaperones BAG3 were greatly up-regulated in skeletal muscle of symptomatic AR113Q male mice. Thus, the entire HSPB8-mediated autophagic machinery seems to be stimulated by the presence of mutant ARpolyQ in the skeletal muscle of SBMA affected mice. These data are consistent with those we reported in muscle of an SBMA-related motoneuron disease, the SOD1-linked familial ALS57,58. Not only HSPB8 and its co-chaperone BAG3, but also the levels of other specific HSPB8 interactors, the muscle specific small heat shock protein HSPB2 and HSPB375 were found to be up-regulated in the same animals. All these modifications were accompanied by an increased BAG3:BAG1 ratio (both at mRNA and protein levels) suggestive of a preferential routing of misfolded proteins to autophagy rather than to UPS49,50, which is known to be a late event in SBMA84. We already demonstrated that the HSPB8-mediated PQC machinery, by facilitating the autophagic process, facilitate the clearance of different motoneuron-associated misfolded proteins (ARpolyQ, mutant SOD1, N-terminal fragment of TDP43)43,44,53. Altogether, the modification of the expression of specific genes encoding for proteins involved in the HSPB8-mediated PQC response here reported, may provide muscle specific biomarkers which could allow to follow the progression of disease in SBMA mice and patients during the validation of therapeutic trials.
When we analysed the spinal cord of the same animals, we did not detect variation in the expression levels of all genes included in this study, with the exception of the relative BAG3:BAG1 ratio, which was found to be increased in symptomatic AR113Q male mice, as compared to all controls tested. This finding suggests preferential activation of the autophagic pathways, instead of the UPS pathway, in the spinal cord of affected mice.
However, differently from the neuropathological features found in ALS, in which several spinal cord cell types are sites of misfolded protein toxicity (e.g.: glial cells, microglia), in SBMA the only cell types affected by the ARpolyQ neurotoxicity are the anterior horn motoneurons, which express very high levels of AR. While this allowed us to estimate that no loss of motoneurons occurs at the symptomatic stages in the mice studied here (see above), the extremely low number of motoneuron present in the spinal cord in relation to all other cell types (interneurons, glial cells microglial cells, etc.) may apparently mask a variation in gene expression. Thus, motoneurons might not be highly affected in these mice at this stage of disease, or even mild variations of expression might have been “diluted” by the large fraction of AR-negative cells (interneurons, glial cells, etc.), thus a population of unresponsive cells to ARpolyQ potential toxicity and in this AR113Q mice line, deficits to motor neurons are not observed until much later.
Therefore, all data here reported proved that the presence of mutant ARpolyQ in muscle cells induces a potent autophagic response, possibly in the attempt to clear cells from toxic misfolded species generated by the mutant protein. HSPB8-mediated PQC machinery seems to be profoundly involved in this response, in agreement with its pro-autophagic activity. These data also confirm the importance of the skeletal muscle in pathological events that are at the basis of SBMA onset and progression16,32,33,85 and support the idea that therapeutical intervention focused on the enhancement/restoration of a correct PQC activity in skeletal muscle might be beneficial in SBMA. In addition, the identification of specific autophagy related molecular markers of skeletal muscle degeneration in SBMA might represent a valuable diagnostic tool for monitoring the disease progression. In this study, we have provided an extensive characterization of the autophagic activation as well as of the HSPB8 machinery in the PQC system response in SBMA muscle.
Further research is needed to establish how muscle responds to ARpolyQ toxicity and the mechanisms capable to mediate autophagy and/or restore a normal autophagic flux in motoneurons or muscle cells may help to design novel and more selective approaches to treat SBMA patients.
Materials and Methods
Generation and maintenance of AR113Q knock-in mice
Animal care and experimental procedures were conducted in accordance with the Italian Institute of Technology and the University of Trento ethical committees and were approved by the Italian Ministry of Health. Generation and genotyping of mice containing androgen receptor with 113 CAG repeats in exon 1 was described previously59,80. Mice genetic background was C57Bl/6J. Females carrying one copy of AR113Q in the X chromosome were crossed with C57Bl/6J mice to maintain the colony. Mice were genotyped by PCR on tail DNA, using REDExtract-N-Amp Tissue PCR kit (Sigma-Aldrich, St Louis, MO, USA) according to manufacturer’s instructions. The mice were housed in filtered cages in a temperature-controlled room with a 12-hour light/12-hour dark cycle with ad libitum access to water and food. Body weight was followed on 5 wt and 20 AR113Q male mice.
Histological analysis on mice spinal cord and muscle sections
Deeply anesthetized mice were transcardially perfused with 4% paraformaldehyde (PFA). Lumbar spinal cords were post-fixed for 24 h in 4% PFA and stored in PBS plus sodium azide. Paraffin embedded lumbar spinal cord was serially sectioned at 20 μm sections, mounted on slides and processed for Nissl staining. Nissl staining on spinal cord was performed on 5 wt and 6 AR113Q male mice on 7 slice/mouse (see below). Motor neurons were identified as cells positive for Nissl staining, with clear nucleus and nucleolus, with a maximum diameter greater than 20 μm leaving 100 μm between slices to avoid counting the same motor neuron. The number of motoneurons/slice in the graph is the result of the average of motoneurons per slice per mouse. Counting was performed in a blinded fashion of all the seven different slices per animal and the total number of motoneurons in these slices and the average number of motoneurons per slice were considered. The mean ± s.e.m. of the number of motor neurons in the spinal cord of 5 control mice and 6 AR113Q mice was calculated.
Muscles were collected immediately after euthanasia, flash-frozen in isopentane precooled in liquid nitrogen and stored at −80 °C until further processing. Frozen muscles were embedded in optimal cutting temperature (OCT) compound (Tissue Tek, Sakura, Torrance, CA) and cross sections (10 μm-thick) were cut with a cryostat (CM1850 UV, Leica Microsystem, Wetzlar, Germany). Cryosections of quadriceps were stained for hematoxylin and eosin (H&E). For hematoxylin and eosin staining, sections were air-dried and incubated in hematoxylin (Sigma-Aldrich) for 3 minutes, then washed in water and incubated in eosin (Roth, Karlsruhe, Germany) for 1 minute. After incubation, sections were washed in water and dehydrated rapidly in 70, 80 and 100% ethanol and xylene. Sections were mounted with di-Nbutyle phthalate in xylene (DPX) mounting media (Sigma-Aldrich). Images were taken using a Nikon Eclipse 90i upright microscope.
Grip strength and Rotarod analyses
The behavior analysis was performed on 5 wt and 15 AR113Q male mice. A Grip Strength Meter (Ugo Basile Instruments) was used to measure forelimb grip strength. The grip strength meter was positioned horizontally and mice were held by the tail and lowered toward the apparatus. Mice were allowed to grasp the smooth metal triangular pull bar with their forelimbs only and then were pulled backward. The force applied to the bar at the moment the grasp was released and recorded as the peak tension. The test was repeated 5 consecutive times within the same session and the highest value from the 5 trials was recorded as the grip strength for that animal. The test was performed weekly from 9 to 24 weeks of age 5 control mice and 15 AR113Q SBMA mice were used in this study. Statistical analysis was performed using a two-tailed paired Student’s t-Test using the PRISM software (GraphPad, San Diego, CA, USA).
For rotarod analysis, mice were placed on a rotating rod using the Ugo Basile Rotarod (Ugo Basile Instruments) at constant speed of 20 rpm for maximum period of 600 seconds. The training of the animals was performed one week before the analysis. Three trials were recorded and the latency to fall was recorded for each trial and the average was used for statistical tests. The test was performed weekly from 9 to 24 weeks of age. 5 control mice and 15 AR113Q mice were used in this study. Statistical analysis was performed using a two-tailed paired Student’s t-Test using the PRISM software (GraphPad, San Diego, CA, USA).
In order to avoid modifications induced by the body weight of the mice in the latency of staying on the grip strength meter or on the rotarod apparatus, both the grip strength and the rotarod tests were also statistically evaluated using body weight as covariate statistical analysis on these tests was also performed using the body weight as covariate.
Filter retardation assay
Spinal cord and quadriceps tissues of 3 control mice and 3 AR113Q mice were collected after euthanasia and store at −80 °C before processing. Tissues were pulverized using pestle and mortar and proteins were extracted using RIPA buffer, containing 150 mM NaCl, 6 mM Na2HPO4, 4 mM NaH2PO4, 2 mM EDTA pH 8, 1% Na-deoxycholate, 0.5% Triton X-100, 0.1% sodium dodecyl sulfate plus protease inhibitor cocktail (Roche). Protein lysates from spinal cord were sonicated and centrifuged at 15.000 rpm for 15 min at 4 °C. Lysates from quadriceps were homogenized with stirrer (Stirrer set RZR 2052, VWR) at 600 rpm, 20 times and flush up and down using syringe of 22G, 25G and 28 G needles and centrifuged at 15.000 rpm for 15 min at 4 °C. Protein concentration was measured using BCA method. 50 μg of total protein were diluted in 100 μl of TBS 1X. The protein solutions were loaded in the dot-blot apparatus (Bio-Rad Laboratories, Hercules, CA, USA) and pass through a 0.2 μm acetate membrane (GE Healthcare Life Sciences, Little Chalfont, UK).
The membranes were blocked for 30 min in 5% BSA or milk in TRIS-buffered saline plus 0.1% Tween-20 (TBS-T) and incubated with anti-androgen receptor antibody (H280, Santa Cruz Biotechnology, Dallas, TX, USA) for 1 h. The membranes were rinsed three times with TBS-T for 10 min, incubated with secondary antibody conjugated with alkaline phosphatase for 30 min and washed three times with TBS-T. Signals were detected using the chemiluminescence reagent ECL (GE Healthcare Life Sciences) and proteins level in each sample was evaluated using Image J 1.48 software.
RNA and protein extraction
Frozen spinal cord and muscle were suspended in TRI-REAGENT (Sigma-Aldrich) and homogenized with Tissue-Lyser (QIAGEN, Valencia, CA, USA) and then RNA was extracted following the TRI-REAGENT Manufacturer’s protocol. After homogenization, 1-bromo-3-chloropropane was added to ensure complete dissociation of nucleoprotein complexes and then the samples were centrifuged to obtain the phase separation. The aqueous phase, containing RNA was isolated and purified. Briefly, aqueous phase was washed with 2-propanol and subsequently with ethanol both to remove the residual phenol and to precipitate RNA, respectively. The RNA pellet was dissolved in RNase-free water (30 μL). The proteins were precipitated from the organic phase with propanol and then the pellets were washed with 0.3 M guanidine hydrochloride/95% ethanol solution (3 times). After the three washes, ethanol was added and the pellets were resuspended using 1% SDS- 8M Urea in Tris HCl pH8.0 solution (100 μL).
Real time PCR
RNA quantification was carried out by absorbance at 260 nm, using NanoDrop 2000 (ThermoFisher Scientific, Waltham, MA, USA). Total RNA (1 μg) was treated with DNAseI (Sigma-Aldrich) and reverse transcribed into cDNA using the High-Capacity cDNA Archive Kit (Life Technologies, Carlsbad, CA, USA).
Primers for real-time PCR were designed using the Primer 3 program and were synthesized by MWG Biotech (Ebersberg, Germany) with the following sequence: mouse AchR: 5’-GTGCTGGGCTCTTTCATCTC -3′ (forward), 5′-TTCTGTGCGCGTTCTCATAC -3′ (reverse); mouse AR: 5′-ATCCCAGTCCCACTTGTGTC-3’(forward), 5′-GGTCTTCTGGGGTGGAAAGT-3′(reverse); mouse Atrogin-1: 5′-GAAGAGAGCAGTATGGGGTCA -3′ (forward), 5′-CTTGAGGGGAAAGTGAGAC-3′ (reverse); mouse ATG10: 5′-TTCACAGCAGATAGGCGATG -3′ (forward), 5′-TGCAGGTCTCGTCACTTCAG-3′ (reverse); mouse ATP6V1B2: 5′-GGCCCACAGAGAATCAGGTA -3′ (forward), 5′-GAGGGTGGGATGTAGGGTTT -3′ (reverse); mouse BAG1: 5′-GAAACACCGTTGTCAGCACT -3′ (forward), 5′-GCTCCACTGTGTCACACTC-3′ (reverse); mouse BAG3: 5′-ATGGACCTGAGCGATCTCA -3′ (forward), 5′-CACGGGGATGGGGATGTA -3′ (reverse); mouse Beclin1: 5′-TGAAATCAATGCTGCCTGGG-3′ (forward), 5′-CCAGAACAGTATAACGGCAACTCC -3′ (reverse); mouse Cullin3: 5′-CACACCAAAGTGCAACATCC -3′ (forward); 5′-CCAACACCAACCTCAGATCC -3′ (reverse); mouse HSPB2: 5′-GCTCAGTGAAGGCAAGTTCC -3′ (forward), 5′-CAGGACATAGGTGCGACAGA-3′(reverse); mouse HSPB3: 5′-CATCATCATCCAGACCTTCG -3′ (forward), 5′-ACTTCCACCACCAAGATTCC-3′ (reverse); mouse HSPB8: 5′-ATACGTGGAAGTTTCAGG CA -3′ (forward), 5′-TCCTTTGACCTAACGCAACC -3′ (reverse); mouse MAP-LC3b: 5′-CGTCCTGGACAAGACCA -3′ (forward), 5′-CCATTCACCAGGAGGAA -3′ (reverse); mouse MyoD: 5′-GGCTACGACACCGCCTACTA -3′ (forward), 5′-GTGGAGATGCGCTCCACTAT -3′ (reverse); mouse Myogenin: 5′-GGGCAATGCACTGGAGTT -3′ (forward), 5′-CACGATGGACGTAAGGGAGT -3′ (reverse); mouse MURF: 5′-ACCTGCTGGTGGAAAACATC-3′(forward), 5’-AGGAGCAAGTAGGCACCTCA-3′(reverse); mouse p62/SQSTM-1: 5′-AGGGAACACAGCAAGCT -3′ (forward), 5′-GCCAAAGTGTCCATGTTTCA -3′ (reverse); mouse Pax7: 5′-GTATGGCCAAACTGCTGTTGAT -3′ (forward), 5′-GGAGTGTTCCCCAAGCTTCA-3′(reverse); mouse PGC1-α: 5′-GGAATGCACCGTAAATCTGC -3′ (forward), 5′-TTCTCAAGAGCAGCGAAAGC-3′ (reverse); mouse TGFβ1: 5′-GAAGGACCTGGGTTGGAAGT -3′ (forward), 5′-CGGGTTGTGTTGGTTGTAGA -3′ (reverse); mouse UBE2Q: 5′-GGAACAGTTGCCTGGATGTT -3′ (forward), 5′-CTGGTGGGTGTAGGCAGAAT -3′ (reverse); mouse GAPDH: 5′-CCAGAACATCATCCCTGCAT -3′ (forward), 5′-CAGTGAGCTTCCCGTTCA -3′ (reverse).
The evaluated efficiency of each set of primers was close to 100%. Real-time PCR was performed using the CFX 96 Real Time System (Bio-Rad Laboratories, Hercules, CA, USA), in a 10 μL total volume, using the iTaq SYBR Green Supermix (Bio-Rad Laboratories) and with 500nmol primers. PCR cycling conditions were as follows: 94 °C for 10 min, 40 cycles at 94 °C for 15 s and 60 °C for 1 min. Melting curve analysis was performed at the end of each PCR assay to control specificity. Data was expressed as Ct values and used for the relative quantification of targets with the ΔΔCt calculation to give N-fold changes in gene expression (2−ΔΔCt). Values were normalized to those of GAPDH.
Western blot analysis
Protein concentration was determined using Pierce 660nM protein assay reagent with Ionic Detergent Compatibility Reagent (IDCR) (Thermo-Fisher Scientific). WB analysis were performed using 10 and 15% SDS-polyacrylamide gel electrophoresis.
20 μg of proteins for each sample were loaded in the gel. The gels were electrotransferred to nitrocellulose membranes using the Trans-turbo Transfer system (Bio-Rad Laboratories). The membranes were incubated with blocking solution (5% non-fat dried milk in TBS-T) for 1h and then incubated o/n with the following primary antibodies: home-made rabbit polyclonal anti-BAG3 (kindly provided by Prof. Serena Carra, Università degli studi di Modena e Reggio Emilia, Italy; 1:6000); rabbit polyclonal anti-BAG1 (Santa Cruz, Dallas, TX, USA, 1:500); home-made rabbit polyclonal anti-HSPB8 (provided by Dr. J. Landry, Quebec, Canada; 1:1000); rabbit polyclonal anti-p62 (Abcam, Cambridge, UK; dilution 1:2000); rabbit polyclonal anti-LC3 (Sigma-Aldrich; dilution 1:1000); mouse monoclonal anti- alpha tubulin (Sigma-Aldrich; dilution 1:2000). After washing, the membranes were incubated for 1 hour with the TBS-T solution containing the peroxidase-conjugated antibody (goat anti-rabbit, goat anti-mouse Santa-Cruz, 1:5000).
The immunoreactive bands were detected with ECL-prime (GE-Healthcare, Maidstone, Uk) and the images were obtained with ChemiDoc XRS system (Bio-Rad Laboratories).
Statistical analysis
Statistical analysis was performed using a one-tailed Student’s t-Test for two group comparisons and two-way ANOVA for three or more groups comparisons using the PRISM software (GraphPad, San Diego, CA, USA). Specific group pair(s) statistical difference was determined by the Bonferroni post-hoc test.
Additional Information
How to cite this article: Rusmini, P. et al. Aberrant Autophagic Response in The Muscle of A Knock-in Mouse Model of Spinal and Bulbar Muscular Atrophy. Sci. Rep. 5, 15174; doi: 10.1038/srep15174 (2015).
References
Kennedy, W. R., Alter, M. & Sung, J. H. Progressive proximal spinal and bulbar muscular atrophy of late onset. A sex-linked recessive trait. Neurology 18, 671–680 (1968).
Sobue, G. et al. X-linked recessive bulbospinal neuronopathy. A clinicopathological study. Brain 112, 209–232 (1989).
Fratta, P. et al. Correlation of clinical and molecular features in spinal bulbar muscular atrophy. Neurology 82, 2077–2084, 10.1212/WNL.0000000000000507 (2014).
Suzuki, K. et al. CAG repeat size correlates to electrophysiological motor and sensory phenotypes in SBMA. Brain 131, 229–239, 10.1093/brain/awm289 (2008).
Atsuta, N. et al. Natural history of spinal and bulbar muscular atrophy (SBMA): a study of 223 Japanese patients. Brain 129, 1446–1455, 10.1093/brain/awl096 (2006).
Adachi, H. et al. Widespread nuclear and cytoplasmic accumulation of mutant androgen receptor in SBMA patients. Brain Res. 128, 659–670 (2005).
Polo, A. et al. Sensory involvement in X-linked spino-bulbar muscular atrophy (Kennedy’s syndrome): an electrophysiological study. J Neurol. 243, 388–392 (1996).
Chua, J. P. & Lieberman, A. P. Pathogenic mechanisms and therapeutic strategies in spinobulbar muscular atrophy. CNS Neurol. Disord. drug targets 12, 1146–1156 (2013).
Yu, Z. et al. Macroautophagy is regulated by the UPR-mediator CHOP and accentuates the phenotype of SBMA mice. PLoS Genet. 7, e1002321, 10.1371/journal.pgen.1002321 (2011).
Chua, J. P. et al. Transcriptional activation of TFEB/ZKSCAN3 target genes underlies enhanced autophagy in spinobulbar muscular atrophy. Hum. Mol. Genet. 23, 1376–1386, 10.1093/hmg/ddt527 (2014).
Mo, K. et al. Microarray analysis of gene expression by skeletal muscle of three mouse models of Kennedy disease/spinal bulbar muscular atrophy. PloS one 5, e12922, 10.1371/journal.pone.0012922 (2010).
Yu, Z., Wang, A. M., Robins, D. M. & Lieberman, A. P. Altered RNA splicing contributes to skeletal muscle pathology in Kennedy disease knock-in mice. Dis. Model. Mech. 2, 500–507, 10.1242/dmm.003301 (2009).
Johansen, J. A. et al. Recovery of function in a myogenic mouse model of spinal bulbar muscular atrophy. Neurobiol. Dis. 34, 113–120, 10.1016/j.nbd.2008.12.009 (2009).
Jordan, C. L. & Lieberman, A. P. Spinal and bulbar muscular atrophy: a motoneuron or muscle disease? Curr. Opin. Pharmacol. 8, 752–758, 10.1016/j.coph.2008.08.006 (2008).
Malena, A. et al. Androgen-dependent impairment of myogenesis in spinal and bulbar muscular atrophy. Acta Neuropathol. 126, 109–121, 10.1007/s00401-013-1122-9 (2013).
Orsucci, D. et al. Myopathic involvement and mitochondrial pathology in Kennedy disease and in other motor neuron diseases. Curr. Mol. Med. 14, 598–602 (2014).
Chua, J. P. et al. Disrupting SUMOylation enhances transcriptional function and ameliorates polyglutamine androgen receptor-mediated disease. J. Clin. Invest. 10.1172/JCI73214 (2015).
Ramzan, F. et al. Distinct Etiological Roles for Myocytes and Motor Neurons in a Mouse Model of Kennedy’s Disease/Spinobulbar Muscular Atrophy. J. Neurosci. 35, 6444–6451, 10.1523/JNEUROSCI.3599-14.2015 (2015).
La Spada, A. R., Wilson, E. M., Lubahn, D. B., Harding, A. E. & Fischbeck, K. H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 352, 77–79 (1991).
Grunseich, C. et al. Early onset and novel features in a spinal and bulbar muscular atrophy patient with a 68 CAG repeat. Neuromuscular Disord 24, 978–981, 10.1016/j.nmd.2014.06.441 (2014).
Fratta, P. et al. Sequencing analysis of the spinal bulbar muscular atrophy CAG expansion reveals absence of repeat interruptions. Neurobiol. Aging 35, 443 e441–443, 10.1016/j.neurobiolaging.2013.07.015 (2014).
Mhatre, A. N. et al. Reduced transcriptional regulatory competence of the androgen receptor in X-linked spinal and bulbar muscular atrophy. Nature Genet 5, 184–188 (1993).
Kazemi-Esfarjani, P., Trifiro, M. A. & Pinsky, L. Evidence for a repressive function of the long polyglutamine tract in the human androgen receptor: possible pathogenetic relevance for the (CAG)n-expanded neuronopathies. Hum. Mol. Genet. 4, 523–527 (1995).
Tut, T. G., Ghadessy, F. J., Trifiro, M. A., Pinsky, L. & L, Y. E. Long polyglutamine tracts in the androgen are associated with reduced trans-activation, impaired sperm production and male infertility. J. Clin. Endocr. Metab. 82, 3777–3782 (1997).
Orr, H. T. & Zoghbi, H. Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30, 575–621, 10.1146/annurev.neuro.29.051605.113042 (2007).
Campisi, J. & d’Adda di Fagagna, F. Cellular senescence: when bad things happen to good cells. Nat.Rev. Mol. Cell Biol. 8, 729–740, 10.1038/nrm2233 (2007).
Grunseich, C., Rinaldi, C. & Fischbeck, K. H. Spinal and bulbar muscular atrophy: pathogenesis and clinical management. Oral Dis. 20, 6–9, 10.1111/odi.12121 (2014).
Poletti, A. The polyglutamine tract of androgen receptor: from functions to dysfunctions in motor neurons. Front. Neuroendocrinol 25, 1–26, 10.1016/j.yfrne.2004.03.001 (2004).
Palazzolo, I. et al. Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron 63, 316–328, 10.1016/j.neuron.2009.07.019 (2009).
Monks, D. A. et al. Overexpression of wild-type androgen receptor in muscle recapitulates polyglutamine disease. Proc Natl. Acad. Sci. USA 104, 18259–18264, 10.1073/pnas.0705501104 (2007).
Huguenard, A. L., Fernando, S. M., Monks, D. A. & Sengelaub, D. R. Overexpression of androgen receptors in target musculature confers androgen sensitivity to motoneuron dendrites. Endocrinology 152, 639–650, 10.1210/en.2010-1197 (2011).
Lieberman, A. P. et al. Peripheral androgen receptor gene suppression rescues disease in mouse models of spinal and bulbar muscular atrophy. Cell Rep. 7, 774–784, 10.1016/j.celrep.2014.02.008 (2014).
Cortes, C. J. et al. Muscle expression of mutant androgen receptor accounts for systemic and motor neuron disease phenotypes in spinal and bulbar muscular atrophy. Neuron 82, 295–307, 10.1016/j.neuron.2014.03.001 (2014).
Galbiati, M. et al. The anabolic/androgenic steroid nandrolone exacerbates gene expression modifications induced by mutant SOD1 in muscles of mice models of amyotrophic lateral sclerosis. Pharmacol. Res. 65, 221–230, 10.1016/j.phrs.2011.12.001 (2012).
Aggarwal, T. et al. Androgens affect muscle, motor neuron and survival in a mouse model of SOD1-related amyotrophic lateral sclerosis. Neurobiol. Aging 35, 1929–1938, 10.1016/j.neurobiolaging.2014.02.004 (2014).
Simeoni, S. et al. Motoneuronal cell death is not correlated with aggregate formation of androgen receptors containing an elongated polyglutamine tract. Hum. Mol. Genet. 9, 133–144 (2000).
Montie, H. L. et al. Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum. Mol. Genet. 18, 1937–1950, 10.1093/hmg/ddp115 (2009).
Montie, H. L., Pestell, R. G. & Merry, D. E. SIRT1 Modulates Aggregation and Toxicity through Deacetylation of the Androgen Receptor in Cell Models of SBMA. J. Neurosci 31, 17425–17436, 10.1523/JNEUROSCI.3958-11.2011 (2011).
Dossena, M. et al. Human adipose-derived mesenchymal stem cells as a new model of spinal and bulbar muscular atrophy. PloS one 9, e112746, 10.1371/journal.pone.0112746 (2014).
Cortes, C. J. et al. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat. Neurosci. 17, 1180–1189, 10.1038/nn.3787 (2014).
Rusmini, P. et al. 17-AAG increases autophagic removal of mutant androgen receptor in spinal and bulbar muscular atrophy. Neurobiol. Dis. 41, 83–95, 10.1016/j.nbd.2010.08.023 (2011).
Carra, S. et al. Alteration of protein folding and degradation in motor neuron diseases: Implications and protective functions of small heat shock proteins. Prog. Neurobiol. 97, 83–100, 10.1016/j.pneurobio.2011.09.009 (2012).
Rusmini, P. et al. Clearance of the mutant androgen receptor in motoneuronal models of spinal and bulbar muscular atrophy. Neurobiol. Aging 34, 2585–2603, 10.1016/j.neurobiolaging.2013.05.026 (2013).
Giorgetti, E. et al. Synergic prodegradative activity of Bicalutamide and trehalose on the mutant Androgen Receptor responsible for Spinal and Bulbar Muscular Atrophy. Hum. Mol. Genet. 10.1093/hmg/ddu419 (2014).
Rusmini, P. et al. Aggregation and proteasome: the case of elongated polyglutamine aggregation in spinal and bulbar muscular atrophy. Neurobiol. Aging 28, 1099–1111, 10.1016/j.neurobiolaging.2006.05.015 (2007).
Rusmini, P. et al. Proteasomal and autophagic degradative activities in spinal and bulbar muscular atrophy. Neurobiol. Dis. 40, 361–369, 10.1016/j.nbd.2010.06.016 (2010).
Dehay, B. et al. Pathogenic lysosomal depletion in Parkinson’s disease. J. Neurosci. 30, 12535–12544, 10.1523/JNEUROSCI.1920-10.2010 (2010).
Settembre, C. & Ballabio, A. TFEB regulates autophagy: An integrated coordination of cellular degradation and recycling processes. Autophagy 7, 17166 [pii] (2011).
Carra, S. et al. Different anti-aggregation and pro-degradative functions of the members of the mammalian sHSP family in neurological disorders. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 368, 20110409, 10.1098/rstb.2011.0409 (2013).
Gamerdinger, M., Carra, S. & Behl, C. Emerging roles of molecular chaperones and co-chaperones in selective autophagy: focus on BAG proteins. J. Mol. Med. 89, 1175–1182, 10.1007/s00109-011-0795-6 (2011).
Fontaine, J. M. et al. Abnormal small heat shock protein interactions involving neuropathy-associated HSP22 (HSPB8) mutants. FASEB J 20, 2168–2170 (2006).
Irobi, J. et al. Mutant HSPB8 causes motor neuron-specific neurite degeneration. Hum. Mol. Genet. 19, 3254–3265, 10.1093/hmg/ddq234 (2010).
Crippa, V. et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS). Hum. Mol. Genet. 19, 3440–3456, 10.1093/hmg/ddq257 (2010).
Crippa, V. et al. A role of small heat shock protein B8 (HspB8) in the autophagic removal of misfolded proteins responsible for neurodegenerative diseases. Autophagy 6, 958–960, 10.4161/auto.6.7.13042 (2010).
Arndt, V. et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 20, 143–148, 10.1016/j.cub.2009.11.022 (2010).
Kwok, A. S. et al. HspB8 mutation causing hereditary distal motor neuropathy impairs lysosomal delivery of autophagosomes. J. Neurochem 119, 1155–1161, 10.1111/j.1471-4159.2011.07521.x (2011).
Crippa, V. et al. Differential autophagy power in the spinal cord and muscle of transgenic ALS mice. Front. Cell. Neurosci. 7, 234, doi:10.3389/fncel.2013.00234 (2013).
Crippa, V. et al. Motoneuronal and muscle-selective removal of ALS-related misfolded proteins. Biochem. Soc. T. 41, 1598–1604, 10.1042/BST20130118 (2013).
Yu, Z. et al. Abnormalities of germ cell maturation and sertoli cell cytoskeleton in androgen receptor 113 CAG knock-in mice reveal toxic effects of the mutant protein. Am. J. Pathol. 168, 195–204, 168/1/195 (2006).
Onesto, E. et al. Muscle cells and motoneurons differentially remove mutant SOD1 causing familial amyotrophic lateral sclerosis. J. Neurochem. 118, 266–280, 10.1111/j.1471-4159.2011.07298.x (2011).
Lieberman, A. P. & Robins, D. M. The androgen receptor’s CAG/glutamine tract in mouse models of neurological disease and cancer. J Alzheimers Dis 14, 247–255 (2008).
Gumucio, J. P. & Mendias, C. L. Atrogin-1, MuRF-1 and sarcopenia. Endocrine 43, 12–21, 10.1007/s12020-012-9751-7 (2013).
Hwang, C. Y. et al. Quantitative proteome analysis of age-related changes in mouse gastrocnemius muscle using mTRAQ. Proteomics 14, 121–132, 10.1002/pmic.201200497 (2014).
Anderica-Romero, A. C., Gonzalez-Herrera, I. G., Santamaria, A. & Pedraza-Chaverri, J. Cullin 3 as a novel target in diverse pathologies. Redox Biol. 1, 366–372, 10.1016/j.redox.2013.07.003 (2013).
Cirak, S. et al. Kelch-like homologue 9 mutation is associated with an early onset autosomal dominant distal myopathy. Brain 133, 2123–2135, 10.1093/brain/awq108 (2010).
Narola, J., Pandey, S. N., Glick, A. & Chen, Y. W. Conditional expression of TGF-beta1 in skeletal muscles causes endomysial fibrosis and myofibers atrophy. PloS one 8, e79356, 10.1371/journal.pone.0079356 (2013).
Eschbach, J. et al. PGC-1alpha is a male-specific disease modifier of human and experimental amyotrophic lateral sclerosis. Hum. Mol. Genet. 10.1093/hmg/ddt202 (2013).
Thau, N. et al. Decreased mRNA expression of PGC-1alpha and PGC-1alpha-regulated factors in the SOD1G93A ALS mouse model and in human sporadic ALS. J. Neuropath. Exp. Neurol. 71, 1064–1074, 10.1097/NEN.0b013e318275df4b (2012).
Pozzi, P. et al. Androgen 5-alpha-reductase type 2 is highly expressed and active in rat spinal cord motor neurones. J. Neuroendocrinol 15, 882–887 (2003).
Marron, T. U. et al. Androgen-induced neurite outgrowth is mediated by neuritin in motor neurones. J. Neurochem 92, 10–20, Doi 10.1111/J.1471-4159.2004.02836.X (2005).
Matsumoto, A. Hormonally induced neuronal plasticity in the adult motoneurons. Brain Res. Bull. 44, 539–547 (1997).
Matsumoto, A., Micevych, P. & Arnold, P. Androgen regulates synaptic input to motoneurones of the adult rat spinal cord. J. Neurosci. 8, 4168–4176 (1988).
Matsumoto, T. et al. The androgen receptor in health and disease. Annu. Rev Physio. 75, 201–224, 10.1146/annurev-physiol-030212-183656 (2013).
Kolb, S. J. et al. Mutant small heat shock protein B3 causes motor neuropathy: utility of a candidate gene approach. Neurology 74, 502–506, 10.1212/WNL.0b013e3181cef84a (2010).
den Engelsman, J. et al. The small heat-shock proteins HSPB2 and HSPB3 form well-defined heterooligomers in a unique 3 to 1 subunit ratio. J. Mol. Biol. 393, 1022–1032, S0022-2836(09)01063-8 10.1016/j.jmb.2009.08.052 (2009).
Sugiyama, Y. et al. Muscle develops a specific form of small heat shock protein complex composed of MKBP/HSPB2 and HSPB3 during myogenic differentiation. J. Biol. Chem. 275, 1095–1104 (2000).
Sun, Y. & MacRae, T. H. Small heat shock proteins: molecular structure and chaperone function. Cell. Mol. Life Sci. 62, 2460–2476, 10.1007/s00018-005-5190-4 (2005).
Gamerdinger, M. et al. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J 28, 889–901, 10.1038/emboj.2009.29 (2009).
Minoia, M. et al. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: Implications for a proteasome-to-autophagy switch. Autophagy 10, 1603–1621 (2014).
Yu, Z. et al. Androgen-dependent pathology demonstrates myopathic contribution to the Kennedy disease phenotype in a mouse knock-in model. J. Clin. Invest. 116, 2663–2672, 10.1172/JCI28773 (2006).
Galbiati, M. et al. ALS-related misfolded protein management in motor neurons and muscle cells. Neurochem. Int. 79, 70–78, 10.1016/j.neuint.2014.10.007 (2014).
Katsuno, M. et al. Disrupted transforming growth factor-beta signaling in spinal and bulbar muscular atrophy. J. Neurosci. 30, 5702–5712, 10.1523/JNEUROSCI.0388-10.2010 (2010).
Flanders, K. C., Ren, R. F. & Lippa, C. F. Transforming growth factor-betas in neurodegenerative disease. Prog. Neurobiol. 54, 71–85 (1998).
Heine, E. M., Berger, T. R., Pluciennik, A., Orr, C. R. & Merry, D. E. Proteasome-Mediated Proteolysis of the Polyglutamine-Expanded Androgen Receptor is a Late Event in SBMA Pathogenesis. J. Biol. Chem. 10.1074/jbc.M114.617894 (2015).
Oki, K. et al. Contractile dysfunction in muscle may underlie androgen-dependent motor dysfunction in SBMA. J. Appl. Phys. jap 00886 02014, 10.1152/japplphysiol.00886.2014 (2015).
Acknowledgements
The following grants are gratefully acknowledged: Fondazione Telethon, Italy (n. GGP14039 to A.P. and n. TCP12013 to M.P); Fondazione Cariplo, Italy (n. 2014-0686 to A.P.); Fondazione AriSLA, Italy (n. ALS_HSPB8 and ALS_Granulopathy to A.P.); Association Française contre les Myopathies (AFM Telethon), France (n. 16406 to A.P. and n. 17793 to M.P.); Provincia Autonoma di Trento (to M.P.); Regione Lombardia (to A.P.); Università degli Studi di Milano e piano di sviluppo UNIMI (linea B to P.R.); Italian Ministry of Health (n. GR-2011-02347198 to V.C. and n. RF-2011-02350097 to M.P.); Muscular Dystrophy Association (n. 92333 to M.P.). U.S. National Institutes of Health (n. R01 NS055746 to A.P.L.).
Author information
Authors and Affiliations
Contributions
P.R., Performed retro transcription of all RNAs samples and RTq-PCR analysis; M.J.P. designed and perfomed behavioral analysis; R.C., design and setting conditions of primers of RTq-PCR of new genes studies; M.E.C., extraction of RNA from spinal cord and muscle tissue; M.M., prepared the figures of the manuscript; M.G., provide expertise on the analysis of gene expression in muscle tissue, based on her related work on ALS; M.P., contributed with M.E.C. in sample extraction and evaluated RNA purity; E.M., performed the statistical analysis and assisted R.C. in the preliminar studies of primers setting; E.G. provide advices on the use of mice and contribute to revision of the manuscript A.P.L., developed the SBMA mice, provided the mice to M.P., actively contributed to the discussion on the experimental approaches and on the results, revised the manuscript; C.M., performed histological analysis in skeletal muscle; A.R., collected tissues from mice; T.A., performed histopathological analysis on spinal cord; M.P., designed experiments related to mice; V.C., contributed in planning the experiments, with discussion of data and revision of the manuscript; A.P., responsible for the design of all experiments, coordination of all activities, written and revised the entire manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Rusmini, P., Polanco, M., Cristofani, R. et al. Aberrant Autophagic Response in The Muscle of A Knock-in Mouse Model of Spinal and Bulbar Muscular Atrophy. Sci Rep 5, 15174 (2015). https://doi.org/10.1038/srep15174
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep15174
This article is cited by
-
Bicalutamide and Trehalose Ameliorate Spinal and Bulbar Muscular Atrophy Pathology in Mice
Neurotherapeutics (2023)
-
Defective excitation-contraction coupling and mitochondrial respiration precede mitochondrial Ca2+ accumulation in spinobulbar muscular atrophy skeletal muscle
Nature Communications (2023)
-
The caspase-6–p62 axis modulates p62 droplets based autophagy in a dominant-negative manner
Cell Death & Differentiation (2022)
-
Targeting Hsp70 facilitated protein quality control for treatment of polyglutamine diseases
Cellular and Molecular Life Sciences (2020)
-
MicroRNAs promote skeletal muscle differentiation of mesodermal iPSC-derived progenitors
Nature Communications (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.