Abstract
RNA interference (RNAi) is a widespread gene-silencing mechanism and is required for heterochromatin assembly in a variety of organisms. The RNA-induced transcriptional silencing complex (RITS), composed of Ago1, Tas3 and Chp1, is a key component of RNAi machinery in fission yeast that connects short interference RNA (siRNA) and heterochromatin formation. However, the process by which RITS is assembled is not well understood. Here, we identified Sgf73, a subunit of the SAGA co-transcriptional complex, is required for pericentromeric heterochromatin silencing and the generation of siRNA. This novel role of Sgf73 is independent of enzymatic activities or structural integrity of SAGA. Instead, Sgf73 is physically associated with Ago1 and Chp1. The interactions among the subunits of the RITS, including those between Tas3 and Chp1, between Chp1 and Ago1, between Ago1 and Tas3, were all impaired by the deletion of sgf73+. Consistently, the recruitment of Ago1 and Chp1 to the pericentromeric region was abolished in sgf73Δ cells. Our study unveils a moonlighting function of a SAGA subunit. It suggests Sgf73 is a novel factor that promotes assembly of RITS and RNAi-mediated heterochromatin formation.
Similar content being viewed by others
Introduction
RNA interference (RNAi) is a gene-silencing mechanism widespread in eukaryotes. RNAi is mediated by small RNAs, including microRNA, Piwi-interacting RNAs and short interfering RNAs (siRNA). Small RNAs bind with Argonaute protein and guide the complex to complementary sequences for repression1. RNAi is well known to regulate post-transcriptional silencing within the cytoplasm2. On the other hand, pilot work in fission yeast3, along with parallel findings in multicellular organisms4,5,6,7, demonstrate that RNAi also triggers chromatin modifications, leading to heterochromatin assembly and transcriptional silencing.
Heterochromatin is a heavily condensed form of chromatin. In fission yeast, constitutive heterochromatin regions are found at telomeres, the silent mating-type locus and pericentromeric repeats8. During S phase, dh and dg repeats from the outer centromeric region are transcribed by RNA polymerase II9. The transcripts are transcribed by an RNA-directed RNA polymerase complex (RDRC) into double-stranded RNA (dsRNA) and are processed into siRNAs by Dicer (Dcr1)10. These siRNAs pass through the Argonaute chaperone complex (ARC) and then are loaded onto the RNA-induced transcriptional silencing complex (RITS)11. RITS is composed of Argonaute (Ago1), Chp1 and Tas3, with Tas3 bridging Ago1 and Chp1 to form a linear architecture12,13. siRNAs guide RITS to the repeats region through base-pairing with the nascent transcripts and the transcripts are sliced by Ago114. Transcripts-bound RITS recruits RDRC to promote further dsRNA and siRNA production15. RITS also recruits histone methyltransferase Clr4 to initiate H3K9 methylation (H3K9me)16. H3K9me is bound by Chp1, which stabilizes the association between RITS and chromatin17. H3K9me serves as a platform to attract other heterochromatin components, including Swi6, Chp2 and SHREC complex, to compact the chromatin further18,19,20. As a key player in the RNAi-mediated heterochromatin assembly, RITS connects transcript cleavage, siRNA production and chromatin modifications21. However, the process by which RITS is assembled and regulated is still not well understood.
The Spt–Ada–Gcn5–acetyltransferase complex (SAGA) is a highly conserved transcriptional co-activator, which is implicated in transcriptional initiation, elongation and mRNA export. Subunits of SAGA can be assembled into modules with distinct activities. These include the structural module (Spt7, Ada1, Spt20), histone acetyltransferase (HAT) module (Gcn5, Ada2, Ada3, Sgf29) and the histone deubiquitylation (DUB) module (Ubp8, Sgf11, Sus1, Sgf73)22,23. Sgf73 serves to anchor the DUB module to SAGA and thus to fully activate the catalytic activity of Ubp824. In addition, Sgf73 is involved in the establishment of a heterochromatin boundary to block the spread of silencing in budding yeast and this requires the HAT activity of SAGA25. Here, we show that Sg73 is required for siRNA production and heterochromatin silencing in fission yeast. Unexpectedly, this novel role of Sgf73 is independent of the enzymatic activity or the structural integrity of SAGA. Instead, Sgf73 is physically associated with RITS. Notably, Sgf73 is required for the integrity of RITS and its recruitment to the heterochromatin region. The results suggest Sgf73-mediated assembly of RITS is critical for RNAi-dependent silencing.
Results and Discussion
Sgf73 is required for heterochromatin silencing
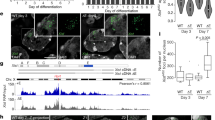
Our previous study showed that sgf73Δ cells are sensitive to thiabendazole (TBZ), a microtubule destabilizing drug26. Hypersensitivity to TBZ could arise from defects in centromeric heterochromatin, because heterochromatin is required to attract cohesin and ensure proper chromosome segregation27. To validate the potential role of Sgf73 in heterochromatin organization, we used a strain in which a ura4+ marker gene was inserted into the outermost (otr) pericentromeric heterochromatin of chromosome 1 (otr1R::ura4+) (Fig. 1a)28. Just as with cells lacking an essential effector of the RNAi machinery (dcr1Δ), deletion of sgf73+ (sgf73Δ) derepressed the ura4+ gene, resulting in poor growth on medium containing counterselective drug 5-fluoroorotic acid (5FOA) and good growth on medium without uracil (Fig. 1b). Consistently, the transcripts from the otr1R::ura4+ and endogenous pericentromeric repeat (dh) and the occupancy of RNA polymerase II at both loci increased substantially in sgf73Δ cells, indicating impaired heterochromatin silencing at the pericentromeric region (Fig. 1c,d). A hallmark of heterochromatin is the presence of histone H3K9 methylation29. Like dcr1Δ cells, sgf73Δ cells exhibited reduced levels of H3K9 dimethylation (H3K9me2) at centromeric repeats (dh) and at the inserted marker ura4+ (Fig. 1e). Thus, we conclude that sgf73+ is essential for maintaining heterochromatin silencing and repressive histone modifications at the pericentromeric region.
Sgf73 is required for heterochromatin silencing at pericentromeric region.
(a) A schematic representation of centromere 1 and the position of a inserted marker gene (otr1R::ura4+). (b) Fivefold serial dilution assay to examine the silencing of otr1R::ura4+. Wild-type (WT) cells with silenced ura4+ grow normally on medium containing 5FOA, while loss of silencing kills cells on 5FOA. (c) RT-PCR analysis of otr1R::ura4+ and pericentromeric repeat (dh) RNA levels relative to a control act1+. The relative level in WT cells was arbitrarily designated as 1. Each column shown in (c) and below represents the mean ± s.d. from three biological repeats. (d,e) ChIP analysis of enrichment of Pol II (d) and H3K9me2 (e) at otr1R::ura4+ and dh relative to fbp1+. Relative enrichment in WT cells was arbitrarily designated as 1.
Sgf73 mediates heterochromatin silencing in a SAGA-independent manner
Sgf73, along with the subunits of the HAT module of the SAGA complex, including Gcn5, Ada2, Ada3 and Sgf29, were shown to act as anti-silencing factors to prevent the spreading of heterochromatin in budding yeast25,30,31,32,33. Unexpectedly, Sgf73 was found to promote silencing in fission yeast in this study (Fig. 1). Therefore, it was of interest to investigate whether the role of Sgf73 in heterochromatin silencing was mediated in the context of the SAGA complex.
SAGA is responsible for the transcription of a broad range of genes. For example, 10% of genes in budding yeast are subjected to regulation by SAGA upon stress response34. However, the mRNA levels of several factors critical for the heterochromatin assembly in sgf73Δ cells were not substantially different with those in WT cells (Fig. 2a, Supplementary Fig. S1). The factors subjected to assay include Ago1, Dcr1, Clr4, Rik1, Swi6, Clr3, Arb1, Arb2, Chp1, Cid12, Raf1, Stc1, Hrr1, Rdp1, Tas3, Clr1, Swi6 and Dsh1. These data are consistent with a previous microarray analysis35. They suggest that Sgf73 regulates heterochromatin silencing without affecting the transcriptions of these essential factors.
The role of Sgf73 in heterochromatin silencing is independent of enzymatic activity and structural integrity of SAGA complex.
(a) RT-PCR analysis of RNA levels of representative factors essential for the heterochromatic silencing. The relative level to a control act1+ in WT cells was arbitrarily designated as 1. Each column shown in (a) and below represents the mean ± s.d. from three biological repeats. (b) Fivefold serial dilution assay to examine the silencing of otr1R::ura4+ in deletion mutants of representative SAGA subunits. (c) RT-PCR analysis of otr1R::ura4+ and pericentromeric repeat (dh) RNA levels relative to a control act1+. (d) ChIP analysis of enrichment of Spt7-3HA at dh, dg, otr1R::ura4+ and mae2+ relative to fbp1+. Relative enrichment in the cells without tagging (no tag) was arbitrarily designated as 1.
To identify the contributions of SAGA to heterochromatin silencing, representative subunits of SAGA were deleted in a strain carrying an otr1R::ura4+ reporter system. In contrast to a severe silencing defect observed in sgf73Δ cells, deletion of other subunit of the UBP module, including Sus1, Sgf11 or the catalytic subunit Ubp8, did not affect silencing of ura4+ (Fig. 2b). Similarly, deletion of subunits of the HAT module, including Sgf29 or the catalytic subunit Gnc5, exhibited no silencing defect (Fig. 2b). Spt7 is a pivotal structural subunit of the SAGA complex. Deletion of Spt7 causes the dissociation of most modules and thus abolishes the activity of the SAGA complex36. As shown in Fig. 2b, albeit suffering a severe growth defect, spt7Δ cells maintained normal silencing of ura4+. Consistently, transcript levels of dh and otr1R::ura4+ in the selected SAGA subunit mutants were kept low as in WT cells, except in the sgf73Δ cells (Fig. 2c). Therefore, these results suggest enzymatic activities and structural integrity of SAGA are not required for the heterochromatin silencing at pericentromeric region.
In a ChIP assay, Spt7 with a C-terminal triple HA tag (Spt7-3HA) was enriched at mae2+, a gene targeted by the SAGA complex35, but not at the centromeric repeat (dh, dg) or inserted ura4+ (Fig. 2d). This result suggests that Spt7, probably in the context of SAGA, is not localized to the pericentromeric heterochromatin region. This is contrast with the enrichment of Sgf73 in heterochromatin shown in Fig. 3 below. Different behaviors between Sgf73 and other subunits of SAGA suggest the role of Sgf73 in the heterochromatin silencing is independent of SAGA. This novel role of Sgf73 might have evolved in fission yeast to cope with different heterochromatin effectors not present in Saccharomyces cerevisiae, such as RNAi37. On the other hand, a SAGA-independent role of Sgf73 is not an exceptional case in fission yeast. Ataxin-7, a human homologue of yeast Sgf73, is associated with microtubules and enhances the stability of microtubule filaments38.
Sgf73 is an essential component of RNAi machinery.
(a) Northern blot analysis of centromeric siRNAs using probes against dg and dh. snoRNA U24 was detected as a loading control. (b) A schematic representation of mating type region and the position of a marker gene inserted into K region (kint2::ura4+). (c) Five-fold serial dilution assay to examine the silencing of kint2::ura4+ in sgf73Δ cells and in strains combining deletion of Sgf73 and deletion of a effector in the RNAi or Atf1/Pcr1 pathway. (d) RT-PCR analysis of kint2::ura4+ RNA levels from the strains in (c). The relative level to a control act1+ in WT cells was arbitrarily designated as 1 Each column shown in (d) and below represents the mean ± s.d. from three biological repeats. (e) Fivefold serial dilution assay to examine the silencing of otr1R::ura4+ in sgf73Δ cells overexpressing dcr1+, ago1+ or clr4+. sgf73Δ cells were transformed with a plasmid overexpressing the indicated gene and transformants were subject to the silencing assay. (f) RT-PCR analysis of dh, dg and otr1R::ura4+ RNA levels relative to a control act1+ in the transformants in (e). (g) ChIP analysis of enrichment of Sgf73-3HA at dh and otr1R::ura4+ relative to fbp1+. Relative enrichment in the cells without tagging (no tag) was arbitrarily designated as 1.
Sgf73 is an essential component of RNAi machinery
Formation and maintenance of centromeric heterochromatin requires RNAi3. As shown in Fig. 1, defective heterochromatin observed in the sgf73Δ cells was similar to that in dcr1Δ cells, suggesting Sgf73 might be involved in the RNAi pathway. Consistent with this idea, siRNAs corresponding to the pericentromeric repeats (dg, dh) were substantially decreased in sgf73Δ cells, albeit not totally abolished as in dcr1Δ cells, while the level of non-coding snoRNA U24 was not affected (Fig. 3a, Supplementary Fig. S2). This suggests Sgf73 is important, but not indispensable for the production of centromeric siRNA.
The involvement of Sgf73 in RNAi machinery was further investigated over the mating type region (mat2P/mat3M), where RNAi and Atf1/Pcr1 act in parallel pathways to nucleate heterochromatin formation39. Deletion of a key component of either pathway, such as Pcr1 or Ago1, did not affect the silencing of a marker ura4+ gene inserted into the K region (kint2::ura4+) (Fig. 3b,c). But disruption of both pathways, as demonstrated by a pcr1Δago1Δ mutant, resulted in the derepression of ura4+ marker and a severe growth defect on 5FOA (Fig. 3c). sgf73Δ and ago1Δ sgf73Δ cells exhibited normal silencing of ura4+, but pcr1Δ sgf73Δ cells were killed on 5FOA due to the derepression of ura4+ (Fig. 3c). Accordingly, the RNA level of ura4+ in pcr1Δ sgf73Δ cells was substantially elevated, as observed in pcr1Δago1Δ cells (Fig. 3d). Overlapping of phenotype of sgf73Δ and ago1Δ cells in heterochromatin silencing over the mating type region strongly suggests Sgf73 acts in the RNAi pathway.
The relationships between Sgf73 and other RNAi components were validated by a complementation assay. The silencing defect of otr1R::ura4+ in sgf73Δ cells was substantially suppressed by overexpressing of dcr1+, ago1+ or clr4+, as shown by the improved growth on 5FOA (Fig. 3e). Accordingly, transcript levels of ura4+ and centromeric repeats (dh, dg) decreased in sgf73Δ cells overexpressing these factors (Fig. 3f). The result suggests Sgf73 is functionally linked with these key players during RNAi-mediated silencing.
Most of the RNAi components, including RITS, RDRP and dicer, are localized at heterochromatin regions, with exceptions of Arb1 and Arb211,13,15,40. To investigate whether Sgf73 mediates heterochromatin silencing in cis, we constructed strains expressing Sgf73 with a C-terminal triple HA tag (Sgf73-3HA). This tag did not affect the gene silencing function of Sgf73 (Supplementary Fig. S3). Compared with a euchromatic locus (fbp1+), Sgf73-3HA was enriched at the endogenous centromeric repeat (dh) and inserted ura4+ marker (otr1R::ura4+) (Fig. 3g). Binding of Sgf73-3HA to heterochromatin decreased substantially in dcr1Δ cells (Fig. 3g), suggesting siRNA is required for the localization of Sgf73 to the heterochromatin region.
Sgf73 is required for the assembly and recruitment of RITS complex
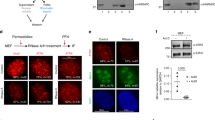
The localization of Sgf73 at the heterochromatin region suggests Sgf73 is physically associated with other RNAi components. As shown in Fig. 4a, Ago1 and Chp1, subunits of RITS complex, were co-purified with Sgf73-3HA in a co-immunoprecipitation assay. This suggests that Sgf73 is associated with RITS. Since Sgf73 was not among the peptides identified by tandem affinity purification using Chp1 as bait15, Sgf73 is unlikely to be a fundamental subunit of the RITS complex. Instead, Sgf73 is perhaps a peripheral component of RITS. Notably, the interactions among the subunits of the RITS, including that between Tas3 and Chp1, between Chp1 and Ago1, were severely impaired by the deletion of sgf73+ (Fig. 4b). Ago1-Tas3 interaction was also disrupted, but to a much lesser extent, in sgf73Δ cells (Fig. 4b). Sgf73 is the first known factor that regulates the integrity of RITS. In contrast, integrity of RITS is not dependent on the production of siRNA or H3K9 methylation, as the Tas3-Ago1-Chp1 composition of RITS is not affected by the deletion of dcr1+or clr4+ 15. A previous study indicates that the physical interaction between Ago1 and Tas3 is required for the recruitment of Ago1 to centromeres for efficient de novo establishment of centromeric heterochromatin41. Accordingly, the enrichments of Ago1 at the centromeric repeat (dh) and inserted ura4+ marker decreased substantially in sgf73Δ cells (Fig. 4c). The enrichment of Chp1 at both loci also decreased, but to a lesser extent comparing to those of Ago1, which might be explained by the tethering of Chp1 with residual H3K9me2 in sgf73Δ cells17. The results suggest that Sgf73 is critical for the recruitment of RITS to the centromeric heterochromatin.
Sgf73 is required for the integrity and recruitment of RITS.
(a) Co-immunoprecipitation (Co-IP) assay to analyze the physical association between Sgf73 and subunits of RITS. Sgf73-3HA immunoprecipitation (IP) was followed by the Western blot (WB) of Ago1. Chp1-3FLAG IP was followed by the WB of Sgf73-3HA. Input and IP samples were run in the same gel. Cropped blots were shown for clarity. Full-length blots are presented in Supplementary Figs S4 and S5. (b) Co-IP assay to analyze the physical association between subunits of RITS in WT and sgf73Δ cells. Tas3-3FLAG IP was followed by the WB of Ago1 and Chp1. Chp1-3FLAG IP was followed by the WB of Ago1. Input and IP samples were run in the same gel. Cropped blots were shown for clarity. Full-length blots are presented in Supplementary Figs S6 and S7. (c) ChIP analysis of enrichment of Ago1 or Chp1 at dh and otr1R::ura4+ relative to fbp1+. Relative enrichment in the WT cells was arbitrarily designated as 1 Each column represents the mean ± s.d. (n = 3) from three biological repeats.
Conclusions
In this study, we revealed an essential role of Sgf73 in RNAi-mediated heterochromatin silencing. Sgf73 is physically associated with RITS and is required for the integrity of RITS. Given the fact that the localizations of Ago1 and Chp1 at heterochromatin were both impaired in sgf73Δ cells, it suggests that RITS need to be fully assembled before it is recruited to the centromeric region to initiate heterochromatin silencing. However, the structural integrity of RITS is not required afterwards to maintain the localization of individual subunits at the centromeric region, as demonstrated by the phenotype of a Tas3 mutant which failed to interact with Ago141. The recruitment of Sgf73 to the centromeric region is probably mediated through the associations with RITS, as the localization of Sgf73 relies on Dcr1 that produces siRNAs to guide RITS.
This study also extends a growing list of the moonlighting functions of SAGA subunits. Besides Sgf73, Spt20, a structural subunit of SAGA, regulates septin ring assembly through physical interactions with septins42. p38IP, a human homologue of Spt20, interacts with mammalian (m)Atg9 (ATG9A) and inhibits the trafficking of mAtg943. Tra1, the largest subunit of SAGA, was identified as a subunit of a novel complex called ASTRA in fission yeast44. Divergently evolved functions of SAGA subunits reflect the capacity of a subunit in a large complex to acquire additional functions to adapt to new environments.
Methods
Yeast strains and plasmids
All the strains used in this study are listed in Supplementary Table S1. Gene deletion and tagging were performed by homologous recombination using a plasmid-based method45. Cells were grown in yeast extract medium with supplements (YES) or Edinburgh minimal glutamate medium minus leucine (EMMG-Leu) medium46. ORF of sgf73+, ago1+, dcr1+ or clr4+ was cloned into pRep41 vector for overexpressing47.
Fivefold serial dilution assay
Exponentially growing cells were collected and adjusted to an A600 of 1.0. Samples were diluted by fivefold for five times. 5 μl dilutions were spotted onto YES or EMMG-Leu medium supplemented with 5FOA (YY12210, Yuanye Biotechnology, Shanghai, China, 1 g/Liter) as indicated. Plates were incubated for 2 or 3d at 32 °C before imaging.
RT-PCR
1 × 108 exponentially growing cells were harvested. Total RNA were extracted using the RiboPure Yeast (AM1926, Life Technologies, Carlsbad, CA, USA) and reverse transcribed into cDNA by using PrimeScript RT (RR037A, Takara, Dalian, China). qPCR was performed using SYBR Premix Ex TaqII (RR820A, Takara) in a LightCycler 480 II Real-Time PCR System (Roche Applied Science, Penzberg, Upper Bavaria, Germany). Primers used are listed in Supplementary Table S2.
ChIP
3 × 108 exponentially growing cells were fixed with 1% formaldehyde for 25 min at 30 °C. After quenching by 250 mM glycine, cells were harvested and washed with Buffer 1 (1 M Tris-HCl (pH 8.0), 167 mM NaCl, 1.2 mM EDTA, 1% TritonX-100, 0.1% Na-deoxycholate). Cells were resuspended in Buffer 1 supplemented with protease inhibitors cocktail (05892970001, Roche Applied Science) and homogenized with a bead-beater (FastPrep-24, MP, California, USA) by glass beads. The cell extract was sonicated for 15 min with a sonicator (Sonics & Materials, Connecticut, USA) and centrifuged. Supernatant was incubated with anti-HA (M20003L, Abmart, Shanghai, China), anti-H3K9me2 (07-441, Millipore, Massachusetts, USA), anti-Ago1 (ab18190, Abcam, Cambs, UK, Abcam), anti-RNA polymerase II 8WG16 (MMS-126R, Covance, New Jersey, USA), anti-FLAG (F1804-200UG, Sigma-Aldrich, St Louis, MO, USA) or anti-Chp1 (ab18181, Abcam) antibody for 4 hour. Samples were subjected to purification by using an EZ-Magna ChIP A Kit (17-408, Millipore). Eluted DNA was subjected to qPCR as described above. Primers used are listed in Supplementary Table S2.
Co-immunoprecipitation
~5 × 1010 exponentially growing cells were harvested. Cells were washed and then resuspended in Buffer 1 supplemented with protease inhibitors as described above. Cells were broken by a high-pressure cell homogenizer (JN-02C, JNBIO, Guangzhou, China) and supernatant were collected after centrifugation. Supernatant was incubated with anti-FLAG M2 Magnetic beads (M8823, Sigma-Aldrich), or with anti-HA antibody and then protein A/G PLUS-agarose beads (sc-2003, Santa Cruz, Dallas, TX, USA) for 6h. Bead-conjugated complexes were washed with Buffer 1, boiled in SDS-gel loading buffer and subjected to Western blot.
Northern blot
Small RNA fractions were prepared from exponentially growing cells using mirVana miRNA Isolation kit (AM1560, Life Technologies). Small RNA was resuspended in 50% formamide and separated on a 15% urea-denaturing poly-acrylamide gel. Samples on the gel were blotted onto a Hybond-N membrane (RPN303N, GE Healthcare, Piscataway, NJ, USA) using a semi-dry electrophoretic transfer cell (170-3912, Bio-Rad, California, USA). 32P labeled oligonucleotide probes complimentary to dh and dg were generated by PCR. Probe against snoU24 was generated by end-labelling. Probes were hybridized to the membrane overnight at 38 °C in a rotating oven. The membrane was then washed twice and exposed to an X-ray film for 3 d at −80 °C. Primers and oligos used are listed in Supplementary Table S2.
Additional Information
How to cite this article: Deng, X. et al. Sgf73, a subunit of SAGA complex, is required for the assembly of RITS complex in fission yeast. Sci. Rep. 5, 14707; doi: 10.1038/srep14707 (2015).
References
Ghildiyal, M. & Zamore, P. D. Small silencing RNAs: an expanding universe. Nature reviews. Genetics 10, 94–108 (2009).
Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nature 457, 413–420 (2009).
Volpe, T. A. et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002).
Baulcombe, D. RNA silencing in plants. Nature 431, 356–363 (2004).
Pal-Bhadra, M. et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303, 669–672 (2004).
Sijen, T. & Plasterk, R. H. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426, 310–314 (2003).
Morris, K. V., Chan, S. W., Jacobsen, S. E. & Looney, D. J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305, 1289–1292 (2004).
Grewal, S. I. & Jia, S. Heterochromatin revisited. Nature reviews. Genetics 8, 35–46 (2007).
Chen, E. S. et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451, 734–737 (2008).
Colmenares, S. U., Buker, S. M., Buhler, M., Dlakic, M. & Moazed, D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Molecular cell 27, 449–461 (2007).
Buker, S. M. et al. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nature structural & molecular biology 14, 200–207 (2007).
Schalch, T., Job, G., Shanker, S., Partridge, J. F. & Joshua-Tor, L. The Chp1-Tas3 core is a multifunctional platform critical for gene silencing by RITS. Nature structural & molecular biology 18, 1351–1357 (2011).
Verdel, A. et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 (2004).
Irvine, D. V. et al. Argonaute slicing is required for heterochromatic silencing and spreading. Science 313, 1134–1137 (2006).
Motamedi, M. R. et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119, 789–802 (2004).
Bayne, E. H. et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140, 666–677 (2010).
Schalch, T. et al. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Molecular cell 34, 36–46 (2009).
Thon, G. & Verhein-Hansen, J. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155, 551–568 (2000).
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001).
Sugiyama, T. et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128, 491–504 (2007).
Creamer, K. M. & Partridge, J. F. RITS-connecting transcription, RNA interference and heterochromatin assembly in fission yeast. Wiley interdisciplinary reviews. RNA 2, 632–646 (2011).
Garcia-Oliver, E., Garcia-Molinero, V. & Rodriguez-Navarro, S. mRNA export and gene expression: the SAGA-TREX-2 connection. Biochim Biophys Acta 1819, 555–565 (2012).
Helmlinger, D. New insights into the SAGA complex from studies of the Tra1 subunit in budding and fission yeast. Transcription 3, 13–18 (2012).
Kohler, A., Zimmerman, E., Schneider, M., Hurt, E. & Zheng, N. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141, 606–617 (2010).
Kamata, K. et al. C-terminus of the Sgf73 subunit of SAGA and SLIK is important for retention in the larger complex and for heterochromatin boundary function. Genes to cells: devoted to molecular & cellular mechanisms 18, 823–837 (2013).
Pan, X. et al. Identification of novel genes involved in DNA damage response by screening a genome-wide Schizosaccharomyces pombe deletion library. BMC Genomics 13, 662 (2012).
Bernard, P. et al. Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542 (2001).
Allshire, R. C., Nimmo, E. R., Ekwall, K., Javerzat, J. P. & Cranston, G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes & development 9, 218–233 (1995).
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000).
Donze, D. & Kamakaka, R. T. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. The EMBO journal 20, 520–531 (2001).
Oki, M., Valenzuela, L., Chiba, T., Ito, T. & Kamakaka, R. T. Barrier proteins remodel and modify chromatin to restrict silenced domains. Molecular and cellular biology 24, 1956–1967 (2004).
Jacobson, S. & Pillus, L. The SAGA subunit Ada2 functions in transcriptional silencing. Molecular and cellular biology 29, 6033–6045 (2009).
Kamata, K. et al. The N-terminus and Tudor domains of Sgf29 are important for its heterochromatin boundary formation function. Journal of biochemistry 155, 159–171 (2014).
Huisinga, K. L. & Pugh, B. F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Molecular cell 13, 573–585 (2004).
Helmlinger, D. et al. Tra1 has specific regulatory roles, rather than global functions, within the SAGA co-activator complex. EMBO J 30, 2843–2852 (2011).
Wu, P. Y. & Winston, F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Molecular and cellular biology 22, 5367–5379 (2002).
Chang, S. S., Zhang, Z. & Liu, Y. RNA interference pathways in fungi: mechanisms and functions. Annual review of microbiology 66, 305–323 (2012).
Nakamura, Y. et al. Ataxin-7 associates with microtubules and stabilizes the cytoskeletal network. Hum Mol Genet 21, 1099–1110 (2012).
Jia, S., Noma, K. & Grewal, S. I. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304, 1971–1976 (2004).
Woolcock, K. J., Gaidatzis, D., Punga, T. & Buhler, M. Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nature structural & molecular biology 18, 94–99 (2011).
Partridge, J. F. et al. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Molecular cell 26, 593–602 (2007).
Lei, B. et al. Septin ring assembly is regulated by Spt20, a structural subunit of the SAGA complex. Journal of cell science 127, 4024–4036 (2014).
Webber, J. L. & Tooze, S. A. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J 29, 27–40 (2010).
Shevchenko, A. et al. Chromatin Central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome biology 9, R167 (2008).
Gregan, J. et al. High-throughput knockout screen in fission yeast. Nature protocols 1, 2457–2464 (2006).
Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods in enzymology 194, 795–823 (1991).
Basi, G., Schmid, E. & Maundrell, K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123, 131–136 (1993).
Acknowledgements
We thank Robin Allshire (The University of Edinburgh, UK) and Shiv Grewal (National Institutes of Health, USA) for providing valuable strains, Rolf Sternglanz (Stony Brook University, USA) for his valuable comments on the manuscript. This work is supported by the National Natural Science Foundation of China (91129730 and 21132004 to HL, 31571287 to YY), the program 863 (2014AA021301 and 2013AA102803B to HL), independent research funding of Fudan University (20520133231 to YY), the Specialized Research Fund for the Doctoral Program of Higher Education (20110071130007 to HL, 20120071120010 to YY) and the Open Fund of State Key Laboratory of Genetic Engineering.
Author information
Authors and Affiliations
Contributions
X.D. designed and performed experiments. H.Z. performed the qPCR analysis. G.Z. performed the Northern blot. W.W. and L.M. assisted the ChIP assay. X.Z. assisted the construction of strains. Y.Y. and H.L. designed and supervised the project. X.D. and Y.Y. wrote the manuscript. All co-authors critically reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Deng, X., Zhou, H., Zhang, G. et al. Sgf73, a subunit of SAGA complex, is required for the assembly of RITS complex in fission yeast. Sci Rep 5, 14707 (2015). https://doi.org/10.1038/srep14707
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep14707
This article is cited by
-
Regulation of centromeric heterochromatin in the cell cycle by phosphorylation of histone H3 tyrosine 41
Current Genetics (2019)
-
Set3 contributes to heterochromatin integrity by promoting transcription of subunits of Clr4-Rik1-Cul4 histone methyltransferase complex in fission yeast
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.