Abstract
Microbial degradation of N-heterocyclic compounds, including xanthine, quinoline, nicotinate and nicotine, frequently requires molybdenum hydroxylases. The intramolecular electron transfer chain of molybdenum hydroxylases consists of a molybdenum cofactor, two distinct [2Fe-2S] clusters and flavin adenine dinucleotide. 3-Succinoylpyridine monooxygenase (Spm), responsible for the transformation from 3-succinoylpyridine to 6-hydroxy-3-succinoylpyridine, is a crucial enzyme in the pyrrolidine pathway of nicotine degradation in Pseudomonas. Our previous work revealed that the heterotrimeric enzyme (SpmA, SpmB and SpmC) requires molybdopterin cytosine dinucleotide as a cofactor for their activities. In this study, we knocked out four genes, including PPS_1556, PPS_2936, PPS_4063 and PPS_4397 and found that a novel gene, PPS_4397 encoding moaE, is necessary for molybdopterin cytosine dinucleotide biosynthesis. Resting cell reactions of the moaE deletion mutant incubated with 3 g l−1 nicotine at 30 °C resulted in accumulation of 3-succinoylpyridine and the strain complemented by the moaE gene regained the ability to convert 3-succinoylpyridine. In addition, reverse transcription-quantitative polymerase chain reaction analysis indicated that the transcriptional levels of the genes of moaE, spmA, and spmC of Pseudomonas putida S16 were distinctly higher when grown in nicotine medium than in glycerol medium.
Similar content being viewed by others
Introduction
Large quantities of tobacco wastes containing high concentration of nicotine are produced during tobacco manufacturing yearly1. Nicotine is well known to be harmful to human health and can cross biological membranes and blood-brain barrier easily2. Microorganisms such as Pseudomonas are important for degrading nicotine and the pyrrolidine pathway of nicotine degradation has been systematically unraveled in Pseudomonas putida strain S163,4,5,6. The pyrrolidine ring is first dehydrogenated to N-methylmyosmine, followed by spontaneous hydrolysis and oxidation to form 3-succinoylpyridine (SP). The molybdenum-dependent enzyme 3-succinoylpyridine monooxygenase (Spm) catalyzes SP to 6-hydroxy-3-succinoylpyridine (HSP) (Fig. 1). This enzyme is composed of three subunits: a large subunit carrying molybdopterin cytosine dinucleotide (Mo-MCD), a middle subunit binding a flavin adenine dinucleotide (FAD) molecule and a small subunit with two [2Fe-2S] clusters3. When the spmABC genes are expressed in Escherichia coli, an inactive Spm is formed that lacks the Mo-MCD cofactor. Instead, the cofactor in E. coli is molybdopterin guanine dinucleotide (MGD), which cannot be integrated into the functional apoprotein Spm3.
3-Succinoylpyridine degradation in P. putida S16.
Spm catalyzes 3-succinoylpyridine (SP) to 6-hydroxy-3-succinoylpyridine (HSP) in the pyrrolidine pathway. Spm has three subunits SpmA, SpmB, SpmC, respectively binding three cofactors Mo-MCD, FAD and [2Fe-2S] clusters. MoaE is crucial for Mo-MCD synthesis and Spm holoenzyme activity.
High-valent molybdenum has been identified as the mononuclear active site of a series of oxidoreductases, on account of its water solubility and general catalytic activity to transfer an oxygen atom either to or from the electron acceptor or donor7,8,9. SpmABC in the pyrrolidine pathway is attributed to typical prokaryotic molybdenum-containing hydroxylases that are found in the biodegradation pathways of some N-heterocyclic compounds10. In Arthrobacter nicotinovorans, nicotine dehydrogenase and ketone dehydrogenase catalyze the formation of pyridine ring-hydroxylated products and Mo-MCD is required for holoenzyme assembly of these two enzymes11,12. Significantly, the biodegradation of isonicotinate by Mycobacterium sp. INA113 and the metabolism of nicotinate by Bacillus niacini14 also have similar hydroxylation steps to generate 2,6-dihydroxy-substituted products, which illustrates the important role of molybdenum cofactor in the metabolism of N-heterocyclic compounds. Therefore, it is crucial to isolate and identify the essential genes of molybdenum cofactor biosynthesis involved in the nicotine degradation of Pseudomonas.
In this study, we disrupted several genes that were selected by mining of the genome sequence of strain P. putida S16 to identify Mo-MCD synthesis-related genes. We found that PPS_4397, encoding moaE, was necessary for Mo-MCD biosynthesis in strain P. putida S16. The moaE deletion mutant strain P. putida S16dmoaE could not grow with nicotine as the sole carbon and nitrogen source. Resting cells of P. putida S16dmoaE incompletely metabolized nicotine and accumulated SP-like P. putida S16dspm3. Here, we identify and characterize the moaE gene required for the synthesis of Mo-MCD in nicotine degradation. The objective of this work was to demonstrate the unknown catabolic genes involved in the nicotine degradation pathway from Pseudomonas.
Results
Identification of genes for Mo-MCD synthesis
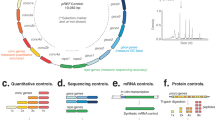
Gene annotation and protein sequence alignment of strain S16 revealed several genes predicted to be potentially responsible for Mo-MCD synthesis. In consideration of the conserved biosynthetic pathway of molybdenum cofactor in all bacteria, gene annotation was quite credible. These predicted genes involved in molybdate uptake, molybdopterin biosynthesis and molybdenum cofactor further modification. Four of these genes (PPS_1556, encoding for molybdenum cofactor biosynthesis protein A; PPS_2936, encoding for molybdopterin-guanine dinucleotide biosynthesis protein A; PPS_4063, encoding for molybdenum cofactor biosynthesis protein; PPS_4397, encoding for molybdopterin biosynthesis MoaE protein) were successfully knocked out by using the suicide plasmid pK18mob, which were confirmed by PCR analysis (Fig. 2B). All the mutants were cultured in Luria-Bertani medium containing 1 g l−1 of nicotine and prepared as resting cells to degrade nicotine. After 8 h, the sample was immediately detected by thin-layer chromatography and only the PPS_4397 gene (moaE) deletion strain could accumulate SP (Fig. 2C).
Plasmid construction and gene disrupted mutants’ confirmation by PCR.
(A) The procedure of plasmid construction. The moaE gene was deleted by single homologous recombination using suicide plasmid pK18mob. (B) PCR analysis of gene disrupted mutants. The former primer is the corresponding upstream gene and the reverse primer is a section sequence of pK18mob. Lane M, marker; lane 1, S16dPPS_1556; lane 2, S16dPPS_2936; lane 3, S16dPPS_4063; lane 4, S16dmoaE; (C) TLC analysis of the supernatant by incubation of 3 g l−1 nicotine and resting cells of gene disrupted mutants at 8h. Resting cells reaction condition is 30 °C and 200 rpm. Lane M, marker containing 1 g l−1 nicotine and 4 g l−1 SP as standards; lane 1, S16dPPS_1556; lane 2, S16dPPS_2936; lane 3, S16dPPS_4063; lane 4, S16dmoaE.
Primary sequence alignment of moaE
The amino acid sequence translated from the moaE gene was compared to known molybdopterin synthase large subunit (MoaE) sequences from seven different species in the National Center of Biotechnology Information data library. Phylogenetic tree construction for the individual proteins was performed using MEGA 4.1 with the neighbor joining method15. The results showed that MoaE from P. putida S16 is closely related to the ortholog from P. mirabilis HI4320 (Fig. 3A). Multiple sequence alignments by Vector NTI revealed that the gene product also contains reported highly conserved sites, including Arg 37, Lys 117 and Lys 12416 (Fig. 3B).
Phylogenetic tree and sequence conservation of MoaE.
(A) Phylogenetic tree of MoaE from eight different strains constructed with the neighbor joining method using MEGA 4.1. (B) Multiply sequence alignment of MoaE from the different eight species using Vector NTI. Identical conservative sites are highlighted in yellow, comparatively conservative sites are highlighted in blue and green.
Cell growth and resting cell reactions of mutant moaE
Cell growth and resting cell reaction evaluations of the moaE gene deletion mutant were performed and monitored by high-performance liquid chromatography (Fig. 4A). The mutant could not grow in the nicotine medium, whereas the wild-type strain S16 grew rapidly in this medium (Fig. 4B). In addition, resting cells of the moaE deletion mutant transformed 3 g l−1 nicotine into SP over 2 h and no subsequent products were detected (Fig. 4C). In order to further verify gene function, the shuttle plasmid pME6032-moaE was transferred into P. putida S16dmoaE. The transformant, P. putida S16dmoaE (pME6032-moaE), recovered the ability to grow with nicotine as the sole carbon and nitrogen source, although the growth rate was slightly slower than that of the wild-type strain (Fig. 4B). Resting cells of strain P. putida S16dmoaE (pME6032-moaE) could also convert SP to downstream metabolites and accumulated only a small amount of SP at 2 h (Fig. 4D). However, resting cells of P. putida S16dmoaE (pME6032-moaEmut) continued to accumulate SP at 8 h (Fig. 4A).
Deletion and complement of moaE gene.
(A) HPLC spectrogram of intermediates produced by reaction of resting cells of P. putida S16, P. putida S16dmoaE, P. putida S16dmoaE (pME6032-moaE) and P. putida S16dmoaE (pME6032-moaEmut) at 30 °C using 3 g l−1 nicotine as the substrate. black: 0 h, red: 2 h, blue: 4 h, green: 8 h. (B) Growth curve of P. putida S16 (■), the moaE gene deletion mutant P. putida S16dmoaE (•) and P. putida S16dmoaE (pME6032-moaE) (▲) with nicotine as the sole carbon and nitrogen source at 30 °C and 200 strokes (rpm). (C) HPLC analysis of nicotine degradation by resting cells of P. putida S16 (■), P. putida S16dmoaE (•), P. putida S16dmoaE (pME6032-moaE) (▲) at 30 °C. (D) HPLC analysis of SP formation by resting cells of P. putida S16 (■), P. putida S16dmoaE (•), P. putida S16dmoaE (pME6032-moaE) (▲) at 30 °C.
RT-qPCR analysis of moaE, spmA, and spmC
RT-qPCR results showed that all target genes related to Spm holoenzyme activity were distinctly up-regulated from P. putida S16 grown in nicotine medium relative to those grown in the glycerol medium. Only a minimal difference was found for expression levels of moaE, which was upregulated 1.8-fold, whereas the mRNAs of spmA and spmC were 4.1- and 2.1-fold upregulated, respectively, with nicotine induction (Fig. 5). These results demonstrated that the transcriptional levels of moaE, spmA, and spmC of P. putida S16 in nicotine medium were distinctly higher than those in the glycerol medium, indicating that these genes are nicotine-induced.
Confirmation of transcriptional levels of differentially expressed proteins that were related to SP degradation.
RT-qPCR analysis of target gene transcripts produced in P. putida S16 grown in the nicotine (black) or glycerol (grey) medium. mRNA expression levels of spmA, spmC and moaE were measured using RT-qPCR and the 2ΔΔCT method, while the 16S rRNA gene was used as the reference gene. Results presented in the bar chart are the means of three parallel experiments and error bars illustrate the standard deviations.
Discussion
Nicotine, the most harmful organic ingredient from tobacco, is a serious threat to human health along with the prosperity of the tobacco industry17. Since P. putida can efficiently degrade nicotine as a sole carbon and nitrogen source18,19,20, analysis of the crucial enzymes in the nicotine metabolic pathway will offer useful guidance for the biological treatment of tobacco wastes and accumulation of valuable intermediate products. Nicotine is classified into N-heterocyclic compounds, which generally require dehydrogenases to catalyze hydroxylation of the aromatic ring21. These dehydrogenases are frequently composed of multiple subunits, one of which binds the molybdopterin dinucleotide cofactor (Moco)22. Moco biosynthesis requires a variety of molybdoenzymes and molybdate23. In Arthrobacter nicotinovorans, MobA, which is responsible for the formation of Moco, was found to be essential for holoenzyme assembly of nicotine dehydrogenase and 6-hydroxypseudooxynicotine dehydrogenase11,24. In addition, SirA2, a sulfur-transferase in Pseudomonas sp. HZN6, was found to be associated with the hydroxylation of SP to HSP25.
Spm shows high similarity to other molybdenum-containing hydroxylases with respect to the amino acid composition of the three subunits. In our previous study, spm genes were expressed in E. coli leading to the formation of inactive Spm, whereas the organic configuration of the molybdenum cofactor in E. coli was found to be MGD3. The available form of Moco is Mo-MCD. Efforts to express and purify Spm in Pseudomonas have been met with little success; thus we could not directly test the content of Mo-MCD in vivo. To confirm that the deficiency of Mo-MCD caused the loss of Spm holoenzyme activity, we searched for genes related to Mo-MCD synthesis in P. putida S16. Here, we provide the first report of the gene moaE, which was found to be crucial for Mo-MCD synthesis and the nicotine metabolic pathway in Pseudomonas. The moaE gene deletion mutant had complete spmABC genes but could not convert SP to HSP, whereas the recombinant strain S16dmoaE(pME6032-moaE) regained the ability to degrade SP. Site-directed mutagenesis suggested that the three conserved sites (Arg 37, Lys 117, Lys 124) were essential for enzyme activity of moaE which was consisted with previous report16. RT-qPCR was conducted to compare the transcriptional levels of moaE, spmA and spmC in P. putida S16 grown in nicotine or glycerol medium. These three genes had relatively low mRNA levels, but appeared to be up-regulated in nicotine-induced P. putida S16. These results strongly suggest that moaE is induced in the presence of nicotine and plays an important role in SP conversion.
In summary, we cloned, sequenced and characterized a novel gene in Pseudomonas that is crucial for the SP degradation involved in nicotine catabolism. The molybdenum cofactor biosynthetic gene was firstly reported in the pyrrolidine pathway of nicotine degradation. Thus these findings will expand our understanding of the molecular mechanisms of nicotine metabolism in Pseudomonas.
Methods
Chemicals and media
L-(-)-Nicotine (≥99% purity) was purchased from Fluka Chemie GmbH (Buchs, Switzerland). 3-Succinoylpyridine (SP) (98%) was obtained from Toronto Research Chemicals (Toronto, Canada). 6-Hydroxy-3-succinoylpyridine was prepared by our own laboratory26. All other chemicals were of analytical grade and commercially available. The “nicotine medium” was a minimal medium containing 13.3 g K2HPO4·3H2O, 4 g KH2PO4, 0.2 g MgSO4·7H2O and 0.5 ml of trace elements solution27. L-(-)-Nicotine was added to this minimal medium after filtration sterilization to a final concentration of 1 g l−1. The “glycerol medium” was a minimal medium added 1% glycerol and 1 g l−1 NH4Cl.
Bacterial strains and culture conditions
P. putida S16 was isolated from tobacco cropping soil28 and cultured at 30 °C in the nicotine medium. E. coli cells and the gene deletion mutants of strain S16 were grown at 37 °C in Luria-Bertani (LB) medium and kanamycin was used for selection at appropriate concentrations27.
DNA manipulation and sequence analysis
Genomic DNA was isolated from strain P. putida S16 by using the Genomic DNA Purification Kit (LifeFeng, China). Purification of PCR products was performed with a Wizard Plus Minipreps DNA purification system (Promega, USA). Isolation of DNA fragments from agarose gels was accomplished with Gel Extraction Kit (Generay, China). Digestions with restriction endonucleases, ligations and transformations were performed according to standard procedures. The sequences were analyzed with Vector NTI DNA analytical software (Invitrogen, USA) and homology searches were performed with the BLAST programs at the National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic tree of MoaE from several different strains was constructed using molecular evolutionary genetics analysis software MEGA 4.1 by neighbor joining (NJ) method and repeated bootstrapping for 1000 times was performed15.
Construction of the moaE gene disrupted mutant P. putida S16dmoaE
The moaE and other related genes in strain S16 were deleted by single homologous recombination (Fig. 2A). An internal fragment of moaE carrying 5′- and 3′ truncations was amplified by PCR and cloned into the multiply cloning site of pK18mob. PCR primer sequences were as follows: moaEint-EcoRI, CGGAATTCCGTTCGACCCGGGAGCCGAGAC and moaEint-HindIII, GCCAAGCTTTGGGTATTTTCCTTCTTCCAG. The recombinant plasmid pK18mob-moaEint was transferred into strain S16 by electroporation according to the following conditions: 0.5–1 μg DNA was added to 100 μl electrocompetent cells of P. putida S16, then the mixture was electroporated at 1.25 kVmm−1, 200 Ω, 25 μF with a Bio-Rad Gene-Pulser Xcell (Bio-Rad Laboratories, Hercules, CA). P. putida S16 exconjugants harboring disrupted genes were isolated on LB medium containing kanamycin and ampicillin. Other genes possibly related to molybdenum cofactor synthesis were disrupted as above. All the mutant strains were tested by PCR to confirm target gene disruption.
Construction of complemented strain
Site-directed mutagenesis was performed by gene synthesis when three sites (Arg 37, Lys 117, Lys 124) were simultaneously replaced by Ala, the mutant of moaE gene was called moaEmut. The moaE gene and moaEmut gene were inserted into pME6032 and then transferred into strain P. putida S16dmoaE by electroporation according to the above methods.
Nicotine bioavailability assay
Strains P. putida S16, S16dmoaE, S16dmoaE (pME6032-moaE) and S16dmoaE (pME6032-moaEmut) were incubated in 100 ml of LB medium containing 1 g l−1 of nicotine and 50 mg l−1 of ampicillin4. After 14 h vigorous shaking at 30 °C, cells were harvested by centrifugation (5,000 × g for 8 min at 4 °C) and washed twice with 50 mM phosphate-buffered saline (pH 7.0). The cells were then resuspended in the double-distilled water at OD600 nm 5 (called resting cells). Nicotine was added to each batch of resting cells with the final concentration of 3 g l−1. The cell suspension was sampled during the reaction, the cells were removed by centrifugation at 12,000 × g for 1 min at 4 °C and the supernatant was collected for analysis.
General analytical techniques
The nicotine and 3-succinoylpyridine (SP) present in the culture medium were detected and quantified by high performed liquid chromatography (HPLC) (Agilent 1200 series) equipped with an Eclipse XDB-C18 column (column size, 250 mm × 4.6 mm; particle size, 5 μm; Agilent, USA). A mixture of methanol −1 mmol H2SO4 (10: 90 v v−1) was used as the mobile phase, at a flow rate of 0.5 ml min−1. Thin-layer chromatography (TLC) analysis was performed on 0.25 mm silica gel (GF254, JYD, Yantai, China) with a mobile phase consisting of chloroform: ethanol: methanol: H2O = 6: 3: 0.4: 0.3 (v v−1). The compounds on the gel were detected under an UV lamp (254 nm).
RNA extraction and RT-qPCR
P. putida S16 was cultured in 5 ml nicotine medium from a single colony on nicotine medium plate. Afterwards, a 1: 50 dilution was inoculated with 50 ml nicotine medium (experimental group) and glycerol medium (control) in three 250 ml flasks. Cells were cultured at 30 °C with vigorous shaking and then harvested at the mid-exponential phase (OD600 nm 0.9). Total RNAs were extracted from about 1 × 109 cells of P. putida S16 by Total RNAprep cell/bacteria kit (Tiangen, China) and reverse transcribed to cDNA using random primers and SuperScript III reverse transcriptase (Invitrogen, USA)3. Quantitative PCR analysis was performed in the CFX96 Real-Time PCR Detection System (Bio-rad, USA) with SYBR GreenI RealMasterMix (Tiangen, China) and 10 fold diluted cDNA as the template. The qPCR primers were designed by Beacon designer and listed in Table 1. Melting curves and annealing temperature were confirmed by temperature gradient PCR. The standard curve for above-mentioned primers was performed with a tenfold dilution of P. putida S16 genomic DNA. The threshold cycle (CT) values for spmA, spmC and moaE gene were measured referring to 16S rRNA gene and the relative transcriptional level was reckoned by the 2ΔΔCT method29.
Nucleotide sequence accession number
The amino acid sequence reported in the present study has been deposited in the GenBank under the accession number AEJ14930.
Additional Information
How to cite this article: Jiang, Y. et al. Functional Identification of a Novel Gene, moaE, for 3-Succinoylpyridine Degradation in Pseudomonas putida S16. Sci. Rep. 5, 13464; doi: 10.1038/srep13464 (2015).
References
Novotny, T. E. & Zhao, F. Consumption and production waste: another externality of tobacco use. Tob. Control. 8, 75–80 (1999).
Hecht, S. S. Tobacco smoke carcinogens and lung cancer. J. Natl. Cancer Inst. 91, 1194–1210 (1999).
Tang, H. et al. Systematic unraveling of the unsolved pathway of nicotine degradation in Pseudomonas. PLoS Genet. 9, e1003923 (2013).
Tang, H. et al. A novel gene, encoding 6-hydroxy-3-succinoylpyridine hydroxylase, involved in nicotine degradation by Pseudomonas putida strain S16. Appl. Environ. Microbiol. 74, 1567–1574 (2008).
Tang, H. et al. A Novel NADH-dependent and FAD-containing hydroxylase is crucial for nicotine degradation by Pseudomonas putida. J. Biol. Chem. 286, 39179–39187 (2011).
Tang, H. et al. Novel nicotine oxidoreductase-encoding gene involved in nicotine degradation by Pseudomonas putida strain S16. Appl. Environ. Microbiol. 75, 772–778 (2009).
Iobbi-Nivol, C. & Leimkühler, S. Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim. Biophys. Acta Bioene. 1827, 1086–1101 (2013).
Ivanov, N. V., Hubalek, F., Trani, M. & Edmondson, D. E. Factors involved in the assembly of a functional molybdopyranopterin center in recombinant Comamonas acidovorans xanthine dehydrogenase. Eur. J. Biochem. 270, 4744–4754 (2003).
Wuebbens, M. M. & Rajagopalan, K. V. Mechanistic and mutational studies of Escherichia coli molybdopterin synthase clarify the final step of molybdopterin biosynthesis. J. Biol. Chem. 278, 14523–14532 (2003).
Hille, R. Molybdenum-containing hydroxylases. Arch. Biochem. Biophys. 433, 107–116 (2005).
Sachelaru, P., Schiltz, E. & Brandsch, R. A functional mobA gene for molybdopterin cytosine dinucleotide cofactor biosynthesis is required for activity and holoenzyme assembly of the heterotrimeric nicotine dehydrogenases of Arthrobacter nicotinovorans. Appl. Environ. Microbiol. 72, 5126–5131 (2006).
Igloi, G. L. & Brandsch, R. Sequence of the 165-kilobase catabolic plasmid pAO1 from Arthrobacter nicotinovorans and identification of a pAO1-dependent nicotine uptake system. J. Bacteriol. 185, 1976–1986 (2003).
Kretzer, A., Frunzke, K. & Andreesen, J. R. Catabolism of isonicotinate by Mycobacterium sp. INA1: extended description of the pathway and purification of the molybdoenzyme isonicotinate dehydrogenase. J. Gen. Microbiol. 139, 2763–2772 (1993).
Nagel, M. & Andreesen, J. R. Purification and characterization of the molybdoenzymes nicotinate dehydrogenase and 6-hydroxynicotinate dehydrogenase from Bacillus niacini. Arch. Microbiol. 154, 605–613 (1990).
Kumar, S., Nei, M., Dudley, J. & Tamura, K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9, 299–306 (2008).
Rudolph, M. J., Wuebbens, M. M., Rajagopalan, K. V. & Schindelin, H. Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat. Struct. Mol. Biol. 8, 42–46 (2001).
Brandsch, R. Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol. 69, 493–498 (2006).
Wada, E. & Yamasaki, K. Mechanism of microbial degradation of nicotine. Science 117, 152–153 (1953).
Li, H. J., Li, X. M., Duan, Y. Q., Zhang, K. Q. & Yang, J. K. Biotransformation of nicotine by microorganism: the case of Pseudomonas spp. Appl. Microbiol. Biotechnol. 86, 11–17 (2010).
Zhong, W. H. et al. Degradation of nicotine in tobacco waste extract by newly isolated Pseudomonas sp. ZUTSKD. Bioresour. Technol. 101, 6935–6941 (2010).
Fetzner, S. Enzymes involved in the aerobic bacterial degradation of N-heteroaromatic compounds: molybdenum hydroxylases and ring-opening 2, 4-dioxygenases. Naturwissenschaften 87, 59–69 (2000).
Bonin, I. et al. Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase. Structure 12, 1425–1435 (2004).
Imperial, J., Hadi, M. & Amy, N. K. Molybdate binding by ModA, the periplasmic component of the Escherichia coli mod molybdate transport system. Biochim. Biophys. Acta Biomem. 1370, 337–346 (1998).
Grether-Beck, S. et al. Structural analysis and molybdenum-dependent expression of the pAO1-encoded nicotine dehydrogenase genes of Arthrobacter nicotinovorans. Mol. Microbiol. 13, 929–936 (1994).
Qiu, J. G. et al. A sirA-like gene, sirA2, is essential for 3-succinoylpyridine metabolism in newly isolated nicotine-degrading Pseudomonas sp. HZN6 strain. Appl. Microbiol. Biotechnol. 92, 1023–1032 (2011).
Yu, H., Tang, H. Z. & Xu, P. Green strategy from waste to value-added-chemical production: efficient biosynthesis of 6-hydroxy-3-succinoyl-pyridine by an engineered biocatalyst. Sci. Rep. 4, 5397 (2014).
Wang, S. N., Liu, Z., Tang, H. Z., Meng, J. & Xu, P. Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype A strain S16. Microbiology 153, 1556–1565 (2007).
Wang, S. N. et al. Biodegradation and detoxification of nicotine in tobacco solid waste by a Pseudomonas sp. Biotechnol. Lett. 26, 1493–1496 (2004).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT methods. Methods 25, 402–408 (2001).
Acknowledgements
This work was supported in part by grants from the Chinese National Science Foundation for Excellent Young Scholars (31422004). This work was supported in part by grants from the Chinese National Natural Science Foundation (31230002, 31470223) and from the National Basic Research Program of China (2013CB733901). We also acknowledge the ‘‘Shanghai Rising-Star Program’’ (13QA1401700) and the ‘‘Chen Xing’’ project from Shanghai Jiaotong University.
Author information
Authors and Affiliations
Contributions
Y.J., G.W. and H.T. conceived and designed the project and experiments. Y.J. and H.T. performed the experiments. Y.J., X.P. and H.T. wrote the paper. Y.J., X.P. and H.T. reviewed the manuscript. All authors reviewed the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Jiang, Y., Tang, H., Wu, G. et al. Functional Identification of a Novel Gene, moaE, for 3-Succinoylpyridine Degradation in Pseudomonas putida S16. Sci Rep 5, 13464 (2015). https://doi.org/10.1038/srep13464
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep13464
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.