Abstract
MicroRNAs (miRNAs) exhibit diverse and important roles in plant growth, development and stress responses and regulate gene expression at the post-transcriptional level. Knowledge about the diversity of miRNAs and their roles in leaf development in celery remains unknown. To elucidate the roles of miRNAs in celery leaf development, we identified leaf development-related miRNAs through high-throughput sequencing. Small RNA libraries were constructed using leaves from three stages (10, 20 and 30 cm) of celery cv.‘Ventura’ and then subjected to high-throughput sequencing and bioinformatics analysis. At Stage 1, Stage 2 and Stage 3 of ‘Ventura’, a total of 333, 329 and 344 conserved miRNAs (belonging to 35, 35 and 32 families, respectively) were identified. A total of 131 miRNAs were identified as novel in ‘Ventura’. Potential miRNA target genes were predicted and annotated using the eggNOG, GO and KEGG databases to explore gene functions. The abundance of five conserved miRNAs and their corresponding potential target genes were validated. Expression profiles of novel potential miRNAs were also detected. Anatomical characteristics of the leaf blades and petioles at three leaf stages were further analyzed. This study contributes to our understanding on the functions and molecular regulatory mechanisms of miRNAs in celery leaf development.
Similar content being viewed by others
Introduction
Celery (Apium graveolens L.) belongs to the Apiaceae family and biennial herbs. Although celery originated from the Mediterranean, it is now cultivated and consumed worldwide1. Celery is rich in carotenoids, flavonoids, volatile oils, vitamins, folic acid and dietary fiber2,3. Celery was initially grown for medicinal use because of its beneficial effects on the digestive and cardiovascular systems4. ‘Ventura’ originated from the United States and was later introduced to China. This cultivar compacts with thick glossy leaves and is well known for its high disease resistance and yield.
In vascular plants, leaves serve important functions in growth and biomass production through photosynthesis and transpiration during development. The leaves (petioles and leaf blades) are the main edible parts in celery. Many complex genetic signals and interactions are involved in cell fate during leaf development. Numerous studies indicated that leaf development is regulated by microRNAs (miRNAs)5,6,7. The overexpression of miR396 can decrease growth-regulating factors (GRFs), which affect cell proliferation in the meristem and developing leaves of Arabidopsis thaliana8. The regulation of lanceolate (LA) by miR319 is required for compound-leaf development in tomatoes6. Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice5.
miRNAs are an abundant new class of non-coding endogenous ~20 nt to 24 nt small RNAs9 that widely exist in animals10, plants11 and some viruses12. This new class of small RNAs has diverse and important roles in plant development, protein degradation, stress responses, signal transduction and metabolism13,14. The mature miRNAs are located in hairpin structures and are generated from primary miRNAs (pri-miRNAs) via at least two RNAseIII-mediated steps15. miRNAs do not directly regulate plant growth and development, but regulate gene expression at the post-transcriptional level9,16.
miRNAs play important roles in leaf development in higher plants. Leaf polarity is regulated by the miR390/ARF pathway17, stomatal patterning is regulated by miR824 via Agamous-like16 (AGL16)18 and leaf shape is regulated by miR396 via GRF19. The CINCINNATA (CIN) gene regulated by miR319/Jaw is required for the differential regulation of cell division during leaf morphogenesis to obtain flat leaves20,21. miR165 and miR166 are essential for controlling leaf patterns (radialization and adaxialization) and for vascular bundle development of leaves by directly regulating homeodomain–leucine zipper (HD-ZIP) transcription factor genes22,23,24.
We previously reported 469 (457 known and 12 novel), 431(418 known and 13 novel) and 346 (341 known and 5 novel) miRNAs of celery varieties, namely, ‘Liuhe Huangxinqin’, ‘Jinnan Shiqin’ and ‘Ventura’ by deep sequencing, respectively25,26. However, knowledge on miRNAs at developmental stages of celery leaves remains unknown. In this study, small RNA libraries were constructed using celery leaves from the three stages and were subjected to deep sequencing and bioinformatics analysis. Five conserved miRNAs associated with celery leaf development and their corresponding potential target genes were identified, meanwhile five novel potential miRNAs with high count were also selected. Their expression profiles were determined at different stages of leaf development by using quantitative real-time PCR (qRT-PCR). The petioles and leaf blades at three stages were also anatomically characterized. Results provide valuable information to elucidate the miRNAs associated with leaf development in celery.
Results
Anatomical characteristics of leaf blades and petioles from ‘Ventura’ at three stages
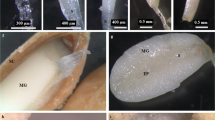
The leaf structure at three development stages (Fig. 1) in ‘Ventura’ was comprehensively investigated using resin-embedded microtome. As shown in Fig. 2, the collenchyma grew stronger and epidermal cells gradually expanded from Stage 1 to Stage 3. The phloem and xylem extensively developed and vascular bundles expanded at the three stages. As shown in Fig. 3, the spongy and palisade mesophyll tissues were gradually and tightly arranged and further developed from Stage 1 to Stage 3. The collenchyma and vascular bundles in the leaf vein further expanded during leaf growth and development.
Structural comparison of ‘Ventura’ leaf blades.
The leaf blade specimens were cut from the main leaf vein and mesophyll tissue near the vein. The specimens were stained with 0.5% methylviolet. A: Mesophyll at Stage 1 × 200; B: Leaf vein at Stage 1 × 200; C: Mesophyll at Stage 2 × 200; D: Leaf vein at Stage 2 × 200; E: Mesophyll at Stage 3 × 200; F: Leaf vein at Stage 3 × 200. Pt: palisade tissues; St: spongy tissues; Ep: epidermis; V: vascular bundles; C: collenchyma.
High-throughput sequencing analysis of small RNAs extracted from celery leaves
To identify miRNAs involved in the development of celery leaves, we performed high-throughput sequencing of small RNA libraries constructed from the three leaf development stages, as follows: Stage 1 (35 d), Stage 2 (50 d) and Stage 3 (65 d). After filtering the low-quality reads, 3’ insert null, poly (A) and reads with <15 or >30 length, we obtained 7,400,022 clean reads at Stage 1, 8,157,893 clean reads at Stage 2 and 13,777,047 clean reads at Stage 3. A total of 29,334,962 clean reads and 7,156,946 unique sequences were obtained. The length of small RNAs varied from 15 nt to 30 nt and the majority of the reads were 21 nt to 24 nt in length (Fig. 4). In the size distribution of the total reads, the 21 nt-long sequence size was the most abundant, followed by 24 nt. Analysis of the size distribution of the unique sequences revealed that the 24-nt small RNAs were the most abundant, followed by 21 nt. This result is consistent with previous reports on other species, such as A. thaliana, cucumber, tomato, trifoliate orange, maize and peanut27,28,29,30,31. Previous studies also reported that 21 nt siRNAs characterize authentic miRNAs and 24 nt siRNAs are associated with repeats and heterochromatic modifications32.
Length distribution of small RNAs in ‘Ventura’.
The reads with a base quality less than 20 were removed and sequences ≤15 nt in length were discarded. Sequences with 15 nt to 30 nt were used for analyses. A: Size distribution of the total small RNA sequences; B: Size distribution of the unique small RNA sequences.
The different types of RNA sequences were further classified by performing BLAST searches against the Rfam database. These sequences included miRNA, rRNA, tRNA, snoRNA, intro and other unannotated reads (Fig. 5). Generally, the distribution of the total small RNA sequences and unique small RNA sequences slightly differed in ‘Ventura’. The distribution of the total and unique sequences at the three stages is shown in Fig. 6.
Distribution of small RNAs among different categories in ‘Ventura’.
The clean reads were annotated as tRNAs, rRNAs, snoRNAs, miRNAs, intro and others based on the Rfam database. A: Distribution of the total small RNA sequences among different categories in ‘Ventura’; B: Distribution of the unique small RNA sequences among different categories in ‘Ventura’.
Identification of conserved and novel miRNAs in celery
To identify conserved miRNAs in ‘Ventura’, we compared all small RNA sequences with currently known miRNAs in the miRNA database miRbase. A total of 333, 329 and 344 unique miRNA sequences (belonging to 35, 35 and 32 miRNA families) were identified in ‘Ventura’ at Stage 1, Stage 2 and Stage 3, respectively (Fig. 7). The base bias on a specific site of miRNAs and the first site of miRNAs with specific lengths are shown in Fig. 8. The majority of these miRNAs started with a 5’-U, which is consistent with typical miRNA sequence patterns33.
To identify novel miRNA sequences, we mapped all unannotated small RNAs onto the celery transcriptome sequence data (SRX326597). Potential pre-miRNAs were searched and hairpin-like structures were predicted. A total of 131 unique sequences were identified as novel miRNAs in ‘Ventura’ (Supplementary File 1). The average minimum free energy value in ‘Ventura’ was −23.63 kcal/mol, which indicated high stability in the hairpin structures.
Prediction and annotation of miRNA putative target genes
miRNA putative target genes were predicted using the psRNA target program and 1,432 potential target genes were identified in ‘Ventura’. To evaluate the completeness of the transcriptome library and the effectiveness of the annotation process, we searched the miRNA target genes against the eggNOG database for functional prediction and classification (Fig. 9)34. A total of 1,974 sequences were assigned to eggNOG classification and they were functionally classified into 26 eggNOG categories. The cluster for ‘function unknown’ was the largest group, followed by ‘general function prediction only’ and ‘transcription’. To describe the miRNA target gene products in terms of their associated biological processes, cellular components and molecular functions, we conducted GO analysis by using Blast2GO (Fig. 10)35. A total of 2,019 target genes were categorized as cellular component, 2,951 as biological process and 800 as molecular function. ‘Metabolic process’ and ‘cellular process’ were the most highly represented groups under the biological process category. For the cellular component category, ‘cell’ and ‘intracellular’ were the most highly represented groups. With regard to molecular function, ‘binding’ was the most highly represented group. The miRNA target genes were assigned based on the KEGG database to special biochemical pathways by using BLASTx (Fig. 11)36. Thirty-four different pathways were found and the most frequently represented pathway was ‘infectious diseases’, followed by ‘immune system’.
Expression profiles of conserved miRNAs and their potential target genes in the petioles and leaf blades at different stages of ‘Ventura’
To investigate the regulatory function and mechanism of the conserved miRNAs and their potential target genes during celery leaf development, the expression of five conserved miRNAs and their potential target genes were validated by qRT-PCR in the petioles and leaf blades at different stages of ‘Ventura’ (Figs 12 & 13). High-throughput sequencing could estimate the expression profiles of miRNA genes. We selected five highly expressed conserved miRNAs from each miRNA family according the high-throughput sequencing data, namely Agr-miR159, Agr-miR164, Agr-miR166, Agr-miR396 and Agr-miR408.
In this study, five conserved miRNAs related to leaf development were expressed in the petioles and leaf blades of ‘Ventura’ at the three stages (Fig. 12). In the petioles, the relative transcript level of Agr-miR408 increased and peaked at Stage 1, decreased at Stage 2 and then continued to decrease and diminish at Stage 3. The relative expression levels of Agr-miR159, Agr-miR164, Agr-miR166 and Agr-miR396 decreased at Stage 2, but increased at Stage 3. The transcript levels were lower at Stage 3 than at Stage 1. In the leaf blades, the transcript levels of Agr-miR159, Agr-miR164, Agr-miR166, Agr-miR396 and Agr-miR408 gradually decreased at the three stages. The transcript levels of Agr-miR159, Agr-miR164, Agr-miR166 and Agr-miR396 at Stage 2 and Stage 3 were low and no significant difference was found at these two latter stages. These results indicated that these five miRNAs are regulated during celery leaf development.
Those five selected miRNAs (Agr-miR159, Agr-miR164, Agr-miR166, Agr-miR396 and Agr-miR408) targeted their corresponding potential genes (Agr-35319 and Agr-26581 gene, Agr-10763 and Agr-25317 gene, Agr-28401 and Agr-20977 gene, Agr-11109 and Agr-29218 gene, Agr-55180 gene), respectively. There is no obvious regularity of the expression levels of their corresponding potential target genes during the 3 stages. By comparing the expression profiles of the five conserved miRNAs and their corresponding potential target genes, we found that the expression of the target genes were independent with miRNAs (Fig. 13).
Expression profiles of novel potential miRNAs in the petioles and leaf blades at different stages of ‘Ventura’
In this study, the expression profiles of five novel potential miRNAs with high count (Agr-miR0056, Agr-miR0002, Agr-miR0005, Agr-miR0046 and Agr-miR0108) were also detected by qRT-PCR (Fig. 14). Significant differences in relative expression levels were measured in the petioles and leaf blades of celery at the 3 stages. The relative expression levels of the five novel potential miRNAs were much higher in the leaf blades than in the petioles of celery. The relative expression levels in the petioles were relatively low and no significant difference was detected among the 3 stages of celery. The relative transcript level in leaf blades was the highest at Stage 1 and relatively low at Stage 2 and Stage 3.
Discussion
miRNAs are important regulators of gene expression at the post-transcriptional level because they repress gene translation. miRNAs play vital roles in plant growth and development and under stressful conditions37,38,39. In this study, miRNAs were identified and characterized using leaves from the three stages of celery cv. ‘Ventura’ through high-throughput sequencing. A total of 333, 329 and 344 known miRNAs (belonging to 35, 35 and 32 families) were detected at Stage 1, Stage 2 and Stage 3, respectively. About 131 novel miRNAs were also identified in ‘Ventura’. The potential target prediction for miRNAs and detailed functional information are important aspects of this study. A total of 1,432 potential target genes were assigned to eggNOG34, GO35 and KEGG classifications40. These results provide useful information for further research on miRNAs that are related to leaf development in celery.
Numerous studies confirmed that small RNAs play important roles in leaf development in higher plants5,41. miRNAs negatively regulate meristem identity, cell division, organ separation, organ polarity and other developmental processes42,43,44. In the present study, five known miRNAs (Agr-miR159, Agr-miR164, Agr-miR166, Agr-miR396 and Agr-miR408) associated with celery leaf development were identified using high-throughput sequencing of small RNAs. Leaf shape is regulated by miR396 via GRF19 and leaf pattern (radialization and adaxialization) is regulated by miR166 via HD-ZIP transcription factors22. miR164 is required to regulate the gene expression of CUC 1 in developing leaf lamina45. miR159/JAW is critical for leaf development and cell division because it targets a subset of TCP transcription factor genes20,46. miR408 is predicted to target LACCASE gene, a regulator of lignin synthesis that is important in xylem and phloem development47,48. Moreover, miR166 targets HD-ZIP transcription factors and is required for leaf vascular bundle development23.
Gene regulation at different stages is closely related to celery leaf development6,7. qRT-PCR not only detects the existence of celery miRNAs, but also their expression trends at different stages. The five miRNA transcripts can be detected in the petioles and leaf blades at the three stages, with the highest expression levels observed at Stage 1. miRNAs also regulate gene expression at the post-transcriptional level by degrading or repressing the translation of targeted miRNAs49. GRF transcription factors mediate the effect on cell proliferation, leaf size and longevity7. HD-ZIP transcription factors also regulate lateral organ polarity and meristem formation50. TCP transcription factor genes play important roles in cell division and signaling pathways that generate different leaf forms51. For example, the edge of leaves may be smooth or jagged and these phenotypes are regulated by the CUC gene family52,53. As shown in Fig. 12, Agr-miR159, Agr-miR164, Agr-miR166 and Agr-miR396 transcripts in the petioles and leaf blades exhibited the highest expression levels at Stage 1. Therefore, we conclude that celery leaf development may be regulated by their target genes (GRF transcription factors, HD-ZIP transcription factors, TCP transcription factors and the CUC gene family), with lower transcript levels at Stage 1 and higher transcript levels at the latter two stages.
Gene regulation at different stages is consistent with structural changes in plants. The transcript levels of Agr-miR408 in the petioles and leaf blades gradually decreased at the three stages. Agr-miR408 and Agr-miR166 transcripts showed the highest levels at Stage 1. LACCASE, a target of miR408, is required for lignin polymerization during vascular development in Arabidopsis54. HD-ZIP transcription factors, a target of miR166, also regulate vascular development55. ATHB15, a member of the HD-ZIP transcription factors, is mainly expressed in vascular tissues and plays important roles in plant vascular system development with expanded phloem and xylem tissues23. Consistent with the anatomical characteristics of the leaf blades and petioles at the three stages, the vascular bundles in the petioles and leaf vein further expanded and the phloem and xylem extensively developed.
Study of the target genes of miRNAs plays an important role in understanding the function and mechanism of miRNAs during leaf development of celery. miRNAs repressed the target gene expression primarily by translation inhibition and degradation of the mRNA. However, it was reported that translation regulation by microRNAs oscillated between repression and activation during the cell cycle10,11,12,13,14. Based on the results of the expression profiles of five conserved miRNAs and their potential target genes in the petioles and leaf blades at different stages of ‘Ventura’, the expression of the corresponding potential target genes was independent from miRNAs. Previous research also found that mutations of some genes compromise miRNA-mediated target gene repression at the protein level, instead of the mRNA. It can be concluded that the five conserved miRNAs regulate the corresponding potential target genes by translation inhibition.
The expression profiles of novel potential miRNAs in the petioles and leaf blades of celery at 3 stages might provide valuable information for the function of these miRNAs during celery leaf development. The data obtained showed potential evidence to support the existence of the novel potential miRNAs in celery. To better elucidate the function and mechanism of these novel potential miRNAs during celery leaf development, more studies need to be performed further.
miRNAs are a class of non-coding endogenous small RNAs that play diverse and important roles in plant growth, development and stress responses. Many experiments demonstrated that miRNAs regulate leaf development, including leaf morphogenesis and polarity and vascular development. miRNAs also regulate the expression of many important genes and most of these genes are transcriptional factors that regulate plant development. In this research, we report a comprehensive study on celery miRNAs by using leaves from the three developmental stages through high-throughput sequencing and high-end bioinformatics methods. A population of small RNAs was characterized as conserved and novel miRNAs. The predicted target genes were subjected to eggNOG, GO and KEGG annotation to infer their functions. The abundance of five conserved miRNAs and their corresponding potential target genes were validated by qRT-PCR in the petioles and leaf blades at different stages of ‘Ventura’. Expression profiles of novel potential miRNAs were also detected. The anatomical characteristics of the leaf blades and petioles at three leaf stages were analyzed by resin embedding with microtomy. Considering that the diversity of miRNAs and their roles in leaf development in celery remain ambiguous, this study provides useful information to elucidate the functions and molecular regulatory mechanisms of miRNAs in celery leaf development.
Methods
Plant materials
Celery cv. ‘Ventura’ seeds were deposited in the State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, China. Celery plants were grown in pots containing peat and vermiculite (2:1,v/v) in a growth chamber with 16 h/8 h at 25 °C/15 °C for day/night, 3 000 lux of light intensity and 75% humidity. Fertilizer and water conditions were strictly controlled.
Three leaf development stages of celery were evaluated and the heights of the celery at Stage 1, Stage 2 and Stage 3 were 10 cm (35 d), 20 cm (50 d) and 30 cm (65 d), respectively (Fig. 1). The leaves were collected at the three stages, immediately frozen in liquid nitrogen and then stored at −80 °C until use. The petioles and leaf blades at the three stages were prepared for resin-embedded sections stained with methylviolet.
Resin embedding with microtomy
Specimens were obtained from the petioles and leaf blades at the three stages by using a razor blade. The petiole specimens were cut from the middle part and the leaf blade specimens were cut from the main leaf vein and mesophyll tissue near the vein. The specimens were prefixed with 2.5% glutaraldehyde at 4 °C overnight. The blocks were washed and dehydrated with an ethanol series of 30% to 100% and then embedded in Spurr low-viscosity embedding medium. Sections (1 μm thick) were cut with a glass knife in a Leica UltracutR microtome (Germany) and then stained with 0.5% methylviolet for 10 min. The sections were visualized and photographed using a CCD camera under Leica DMLS light microscope.
RNA extraction, small RNA library construction and sequencing
Small RNAs were extracted from the leaves at the three stages by using TRIZOL method to construct small RNA libraries. Three small RNA samples were sequenced with an Illumina Hi-seq 2000 platform (Shanghai Personal Biotechnology Co., Ltd.).
Bioinformatics analysis of small RNA sequences
Sequencing reads were generated from the three constructed small RNA libraries. The raw sequences were subjected to bioinformatics analysis to process and identify the sequences representing conserved and novel miRNAs56. The low-quality sequences (reads with a base quality less than 20) were removed and all sequences ≤15 nt in length were discarded. Sequences with 15 nt to 30 nt were used for further analyses. The clean reads were annotated as tRNAs, rRNAs, snoRNAs, miRNAs, intro and other unannotated reads according to the Rfam (11.0 release) database. The unique sequences were used for BLASTN search against the miRNA database (miRBase16.0) and the perfectly matched sequences were identified as conserved miRNAs. Potential novel sequences were identified through alignment with celery transcriptome data.
Prediction of potential miRNA target genes and functional annotation
The potential target genes of celery miRNAs were predicted using the psRNA target program (http://plantgrn.noble.org/psRNATarget/?function=3). The rules used for target prediction were based on those suggested by Schwab et al.57,58 and Allen et al.57,58. Detailed functional information is important to elucidate the overall expression profiles of potential target genes. Numerous target sequences were assigned to various eggNOG, GO and KEGG classifications34,35,59.
RNA extraction and qRT-PCR analysis
Total RNA sequences were extracted using an RNAsimple Total RNA Kit (Tiangen-bio, Beijing, China) according to the manufacturer’s instructions. RNA quality and purity were assessed with OD260/280 ratio and determined using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE). Each sample (10 μg) was reverse transcribed into cDNA using the Prime Script RT reagent Kit (TaKaRa, Dalian, China) and Small RNAs were reverse transcribed into cDNA by using the PrimeScriptTM miRNA qPCR Starter Kit Ver.2.0 (TaKaRa, Dalian, China).
qRT-PCR was performed using the ABI 7300 Real-Time PCR System in accordance to the manufacturer’s instructions (SYBR Premix Ex TaqTM and SYBR Green, TaKaRa, Dalian, China). PCR reactions of miRNAs were performed as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. PCR reactions of potential target genes were performed at 94 °C for 30 s, followed by 40 cycles of 94 °C for 10 s and 58 °C for 20 s. A melting curve (61 cycles at 65 °C for 10 s) was generated to check amplification specificity. Each reaction presented three biological repeats and 5S rRNA and Agactin gene were used as internal controls respectively. The primers used for qRT-PCR are listed in Tables 1, 2, 3. Ct values were represented by the mean values of three independent replicates and the relative gene expression levels were calculated using the △△Ct method60. Standard errors of mean among the replicates were also calculated. Primers were synthesized by GenScript (Nanjing, China).
Additional Information
How to cite this article: Jia, X.-L. et al. High-throughput sequencing of small RNAs and anatomical characteristics associated with leaf development in celery. Sci. Rep. 5, 11093; doi: 10.1038/srep11093 (2015).
References
Lin, L. Z., Lu, S. & Harnly, J. M. Detection and quantification of glycosylated flavonoid malonates in celery, Chinese celery and celery seed by LC-DAD-ESI/MS. J Agric Food Chem 55, 1321–1326 (2007).
Momin, R. A. & Nair, M. G. Antioxidant, cyclooxygenase and topoisomerase inhibitory compounds from Apium graveolens Linn. seeds. Phytomedicine 9, 312–318 (2002).
Kulshrestha, V. K., Singh, N., Saxena, R. C. & Kohli, R. P. A study of central pharmacological activity of alkaloid fraction of Apium graveolens Linn. Indian J Med Res 58, 99–102 (1970).
Raffo, A. et al. Internal quality of fresh and cold stored celery petioles described by sensory profile, chemical and instrumental measurements. Eur Food Res Technol 222, 590–599 (2006).
Xie, K. et al. Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol 158, 1382–1394 (2012).
Ori, N. et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39, 787–791 (2007).
Debernardi, J. M. et al. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J 79, 413–426 (2014).
Mecchia, M. A., Debernardi, J. M., Rodriguez, R. E., Schommer, C. & Palatnik, J. F. MicroRNA miR396 and RDR6 synergistically regulate leaf development. Mech Dev 130, 2–13 (2013).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism and function. Cell 116, 281–297 (2004).
Ambros, V. The functions of animal microRNAs. Nature 431, 350–355 (2004).
Sun, G. L. MicroRNAs and their diverse functions in plants. Plant Mol Biol 80, 17–36 (2012).
Cullen, B. R. Viruses and microRNAs. Nat Genet 38, S25–S30 (2006).
Zhang, B. H., Wang, Q. L. & Pan, X. P. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol 210, 279–289 (2007).
Zhang, B. H., Pan, X. P., Cobb, G. P. & Anderson, T. A. Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289, 3–16 (2006).
Voinnet, O. Origin, biogenesis and activity of plant microRNAs. Cell 136, 669–687 (2009).
Carrington, J. C. & Ambros, V. Role of microRNAs in plant and animal development. Science 301, 336–338 (2003).
Schwab, R., Ossowski, S., Riester, M., Warthmann, N. & Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133 (2006).
Kutter, C., Schöb, H., Stadler, M., Meins, F. Jr. & Si-Ammour, A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 19, 2417–2429 (2007).
Liu, D. M., Song, Y., Chen, Z. X. & Yu, D. Q. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol Plant 136, 223–236 (2009).
Palatnik, J. F. et al. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263 (2003).
Yoo, B. C. et al. A systemic small RNA signaling system in plants. Plant Cell 16, 1979–2000 (2004).
Juarez, M. T., Kui, J. S., Thomas, J., Heller, B. A. & Timmermans, M. C. MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88 (2004).
Kim, J. et al. MicroRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J 42, 84–94 (2005).
Zhou, G. K., Kubo, M., Zhong, R. Q., Demura, T. & Ye, Z. H. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol 48, 391–404 (2007).
Li, M. Y. et al. High throughput sequencing of two celery varieties small RNAs identifies microRNAs involved in temperature stress response. BMC Genomics 15, 242 (2014).
Jiang, Q. et al. High-throughput analysis of small RNAs and characterization of novel microRNAs affected by abiotic stress in a local celery cultivar. Sci Hortic-amsterdam 169, 36–43 (2014).
Rajagopalan, R., Vaucheret, H., Trejo, J. & Bartel, D. P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20, 3407–3425 (2006).
Martínez, G., Forment, J., Llave, C., Pallás, V. & Gómez, G. High-throughput sequencing, characterization and detection of new and conserved cucumber miRNAs. PLoS One 6, e19523 (2011).
Song, C. N. et al. Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics 11, 431 (2010).
Li, D. et al. Deep sequencing of maize small RNAs reveals a diverse set of microRNA in dry and imbibed seeds. PLoS One 8, e55107 (2013).
Chi, X. Y. et al. Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing. PLoS One 6, e27530 (2011).
Pontes, O., Costa-Nunes, P., Vithayathil, P. & Pikaard, C. S. RNA polymerase V functions in Arabidopsis interphase heterochromatin organization independently of the 24-nt siRNA-directed DNA methylation pathway. Mol Plant 2, 700–710 (2009).
Wang, X. & Liu, X. S. Systematic curation of miRBase annotation using integrated small RNA high-throughput sequencing data for C. elegans and Drosophila. Front Genet 2, 25–25 (2011).
Powell, S. et al. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res 40, D284–D289 (2012).
Park, J. C., Kim, T. E. & Park, J. Monitoring the evolutionary aspect of the Gene Ontology to enhance predictability and usability. BMC Bioinformatics 9, S7 (2008).
Wrzodek, C., Büchel, F., Ruff, M., Dräger, A. & Zell, A. Precise generation of systems biology models from KEGG pathways. BMC Syst Biol 7, 15 (2013).
Shen, J. Q., Xie, K. B. & Xiong, L. Z. Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol Genet Genomics 284, 477–488 (2010).
Gleeson, M., Constantin, M., Carroll, B. J. & Mitter, N. MicroRNAs as regulators of adventitious root development. J Plant Biochem Biot 23, 339–347 (2014).
Bertolini, E. et al. Addressing the role of microRNAs in reprogramming leaf growth during drought stress in Brachypodium distachyon. Mol Plant 6, 423–443 (2013).
Kanehisa, M. et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42, D199–D205 (2014).
Mao, Y. F. et al. MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol 164, 710–720 (2014).
Wong, C. E. et al. MicroRNAs in the shoot apical meristem of soybean. J Exp Bot 62, 2495–2506 (2011).
Wang, L. et al. miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J ExpBot 62, 761–773 (2011).
Rodriguez, R. E. et al. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112 (2010).
Sieber, P., Wellmer, F., Gheyselinck, J., Riechmann, J. L. & Meyerowitz, E. M. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134, 1051–1060 (2007).
Li, Y., Li, C. Q., Ding, G. H. & Jin, Y. X. Evolution of MIR159/319 microRNA genes and their post-transcriptional regulatory link to siRNA pathways. BMC Evol Biol 11, 122 (2011).
Lu, S., Li, L., Yi, X., Joshi, C. P. & Chiang, V. L. Differential expression of three eucalyptus secondary cell wall-related cellulose synthase genes in response to tension stress. J Exp Bot 59, 681–695 (2008).
Lu, S., Yang, C. & Chiang, V. L. Conservation and diversity of microRNA-associated copper-regulatory networks in Populus trichocarpa. J Integr Plant Biol 53, 879–891 (2011).
Axtell, M. J. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64, 137–159 (2013).
Zhong, R. Q. & Ye, Z. H. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol 45, 369–385 (2004).
Koyama, T., Mitsuda, N., Seki, M., Shinozaki, K. & Ohme-Takagi, M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22, 3574–3588 (2010).
Nikovics, K. et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18, 2929–2945 (2006).
Berger, Y. et al. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136, 823–832 (2009).
Zhao, Q. et al. LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25, 3976–3987 (2013).
Figueiredo, E., Teixeira, R. & Canhoto, J. Role of several vascular system genes under the control of two class III HD-ZIP promoters in Arabidopsis thaliana. J Biotechnol 150, S488–S489 (2010).
Sunkar, R. & Jagadeeswaran, G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol 8, 37 (2008).
Allen, E., Xie, Z. X., Gustafson, A. M. & Carrington, J. C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 (2005).
Schwab, R. et al. Specific effects of microRNAs on the plant transcriptome. Dev Cell 8, 517–527 (2005).
Altman, T., Travers, M., Kothari, A., Caspi, R. & Karp, P. D. A systematic comparison of the MetaCyc and KEGG pathway databases. BMC Bioinformatics 14, 112 (2013).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45 (2001).
Acknowledgements
The research was supported by the National Natural Science Foundation of China (31272175); New Century Excellent Talents in University (NCET-11-0670); Jiangsu Natural Science Foundation (BK20130027); Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Contributions
X.A.S. and X. L. J. initiated and designed the research. X.L.J., M.Y.L., Q.J., Z.S.X., F.W. and A.S.X. performed the experiments. X.L.J., M.Y.L. and Q.J. analyzed the data. A.S.X. contributed reagents/materials/analysis tools. X.L.J. wrote the paper. X.L.J. and A.S.X. revised the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Jia, XL., Li, MY., Jiang, Q. et al. High-throughput sequencing of small RNAs and anatomical characteristics associated with leaf development in celery. Sci Rep 5, 11093 (2015). https://doi.org/10.1038/srep11093
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep11093
This article is cited by
-
Cucumber mosaic virus-induced gene and microRNA silencing in water dropwort (Oenanthe javanica (Blume) DC)
Plant Methods (2024)
-
Whole transcriptome analysis and construction of a ceRNA regulatory network related to leaf and petiole development in Chinese cabbage (Brassica campestris L. ssp. pekinensis)
BMC Genomics (2023)
-
Unraveling MicroRNA Mediated Gene Regulation in Centella asiatica (L.) Urb. by high-Throughput Sequencing Based Small-RNA Profiling
Tropical Plant Biology (2023)
-
Identification and Expression Analysis of the SBP-box Gene Family Related to Abiotic Stress in Tea Plant (Camellia sinensis (L.) O. Kuntze)
Plant Molecular Biology Reporter (2022)
-
Effect of Elevated CO2 on Ascorbate Accumulation and the Expression Levels of Genes Involved in Ascorbate Metabolism in Celery
Journal of Plant Growth Regulation (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.