Abstract
MTHFR C677T polymorphism has been indicated to be a risk factor for cancers, but its association with head and neck cancer (HNC) risk remains inconclusive. In the present study, we aimed to get a more precise estimation by performing a quantitative meta-analysis. Published papers up to Jun 2014 was searched and screened. Necessary information was rigorously extracted for data pooling and analyzing and then, subgroup analyses on ethnicity, source of controls, sample size, tumor type, smoking and drinking status were also carried out. As a result, twenty-three case-control studies including 14298 subjects were included. The overall data failed to reveal a significant association between MTHFR C677T polymorphism and HNC risk (homozygote comparison model: OR = 1.16; 95%CI = 0.93-1.45; dominant model: OR = 1.05; 95%CI = 0.90-1.21; recessive model: OR = 1.14; 95%CI = 0.93-1.38). However, in the subgroup analysis about drinking status, increase risk was shown in the heavy drinking subgroup (TT vs CC: OR = 3.11; 95%CI = 1.52-3.02). In conclusion, the results of the present study suggest that Homozygous TT alleles of MTHFR C677T polymorphism might be a risk factor for HNC among individuals who have a heavy drinking history. Further studies are needed to get a more definitive conclusion.
Similar content being viewed by others
Introduction
Head and neck carcinoma (HNC), the sixth most frequent kind of cancer worldwide, is a group of biologically similar cancers that originate from head and neck regions such as oral cavity, pharyngeal cavity and larynx1. Previous reports showed that life-style factors such as smoking, drinking, betel quid chewing, papilloma virus infection and exposure to toxic substances are possible etiological risk factors for HNC2,3. Nevertheless, though many individuals are exposed to these external factors, HNC develops only in a small proportion of the exposed people, indicating that intrinsic factors such as genetic polymorphism might play critical roles in its carcinogenic mechanisms.
Generally, folate plays a fundamental role of providing methyl groups for de novo deoxynucleoside synthesis and for intracellular methylation reactions in humans4. Low folate levels may result in uracil misincorporation during DNA synthesis, impaired DNA repair and chromosomal damage5. Several key enzymes were involved in the folate metabolism and their functions may have an effect on folate levels and DNA methylation. Methylenetetrahydrofolate reductase (MTHFR), a key enzyme for intracellular folate hemeostasis and metabolism, catalyses the irreversible conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate that is the primary circulating form of folate, providing methyl groups for the methylation of homocysteine to methionine6,7. A common variation of MTHFR (rs1801133), C677T in exon 4 (Ala222Val), may be implicated in tumorigenesis with alteration of MTHFR enzyme activity8. The homozygous genotype 677TT has been indicated to have only 30% of the MTHFR enzyme activity of the 677CC wild-type genotype9. Therefore, alteration of the enzyme activity resulted from the polymorphism has been thought to be associated with cancer progression and development10.
Published data on the association of MTHFR polymorphism with HNC have generated inconclusive results. Clarifying this association may help us better understand the possible risk of HNC and therefore contribute to its prevention. Previously, we assessed the relationship between MTHFR polymorphism and oral cancer risk11. Recently, several original studies have been reported. Thus, in the present study, we performed an updated meta-analysis including the recent investigations that were conducted on head and neck cancers.
Materials and methods
Literature search strategy
The meta-analysis was presented according the PRISMA-MOOSE statement. We carried out a search in the internet covering well-known biomedical databases such as Medline, EMBASE and CNKI without a language limitation. Papers published up to Jun 1, 2014 were included. The literature selection was performed from Jun 1, 2014 to Jun 30, 2014. The following keywords were used for searching: methylenetetrahydrofolate reductase, MTHFR, head and neck, oral, pharynx, larynx, thyroid, mouth, neoplasm, tumor, cancer, variation and polymorphism. All searched studies were retrieved and the bibliographies were checked for other relevant publications. Review articles and bibliographies of other relevant studies identified were hand searched to find additional eligible studies.
Inclusion criteria
The following criteria were used for the literature selection: First, studies should concern the association of MTHFR C677T polymorphism with HNC risk; second, studies should be case-control or cohort designed; third, papers must offer the size of the sample, odds ratios (ORs) and their 95% confidence intervals (CIs), the genetic distribution or the information that can help infer the results. After deliberate searching, we reviewed all papers in accordance with the criteria defined above for further analysis.
Data extraction
Information was carefully extracted from all eligible papers by two of the authors (XZ and JS) independently according to the inclusion criteria mentioned above. If their results were conflicting, an agreement was reached following a discussion. If a consensus could not be reached, another author (DL) was consulted to resolve the dispute and then a final decision was made by the majority of the votes. The extracted data were entered into a database.
Statistical analysis
The odds ratio (OR) of MTHFR C677T polymorphisms and HNC risk was estimated for each study. The pooled ORs were performed for a homozygote comparison model (TT versus CC), a dominant model (TT + CT versus CC) and a recessive model (TT versus CT + CC), respectively. For detection of any possible sample size biases, the OR and its 95% confidence interval (CI) to each study was plotted against the number of participants respectively. For assessment of the heterogeneity between studies, two indexes were calculated. One was I2 metric, with I2 = 0-25% indicating no heterogeneity, I2 = 25-50% indicating moderate heterogeneity and I2 > 50% indicating large heterogeneity12. The other index was a Chi-square based Q statistic test. If the result of the Q-test was P > 0.1 (indicating no heterogeneity), ORs were pooled according to the fixed-effect model (Mantel-Haenszel), Otherwise, the random-effect model (DerSimonian and laird) was used. The significance of the pooled ORs was determined by Z-test. The Hardy-Weinberg equilibrium (HWE) was assessed via Fisher’s exact test. Publication bias was assessed by visual inspection of funnel plots13, in which the standard error of log (OR) of each study was plotted against its log (OR). The symmetry of the funnel plot was further evaluated by Egger’s linear regression test14. An asymmetric plot indicates a possible publication bias. Statistical analysis was conducted by using the STATA 11.0 software (Stata Corporation, Texas).
Results
Study characteristics
Publications relevant to the key words were retrieved and screened originally. A total of ninety-seven studies were searched and screened for retrieval, of which sixty-seven irrelevant studies were excluded. Then, three review articles15,16,17 were excluded. Next, three studies that were not case-control designed18,19,20 (also not cohort designed) were discarded. Thus, a total of twenty-four papers were included for data extraction. Nevertheless, one duplicate publication21 was further excluded. Lastly, twenty-three case-control studies22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44 were selected (Fig. 1).
All the included studies were written in English except for one study in Chinese30. We established a database according to the extracted information from each article. The relevant information was listed in Table 1. According to the lists, the first author and the number and characteristics of cases and controls for each study as well as other necessary information were presented. As shown in this table, the selected studies involved cancers originated from oral cavity, pharynx, larynx and thyroid.
In the included studies, there were eight groups of Caucasians23,27,29,33,38,41,42,43, eight of Asians22,30,32,35,37,39,40,44 and seven of mixed populations24,25,26,28,31,34,36. Information about smoking could be extracted from five studies22,25,30,39,40 and data regarding drinking status were also available from five studies23,25,37,38,39. The distributions of MTHFR genotype of the included studies were also presented (Table 2). Of note, the data about CT and TT in the study by Hsiung et al.26 were combined as TT + CT and therefore, the relevant data were only included in the dominant model for the whole evaluation. The genetic distributions of the control groups were consistent with HWE except for those in four studies24,33,41,42.
Meta-analysis results
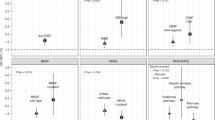
We evaluated the heterogeneity for the homozygote comparison model (TT versus CC), dominant model (TT + CT versus CC) and recessive model (TT versus CT + CC), respectively. As shown in Table 3, the heterogeneity for the overall data was significant in the three models, respectively, because the P value was less than 0.1 for Q-tests and I-squared values indicated more than moderate heterogeneity. Therefore, the random-effect models were used in the present meta-analysis.
Table 3 lists the main results of the meta-analysis. The overall data in the homozygote comparison model (OR = 1.16; 95%CI = 0.93-1.45), the dominant (OR = 1.05; 95%CI = 0.90-1.21) and the recessive models (OR = 1.14; 95%CI = 0.93-1.38) failed to reveal a marked association between MTHFR C677T polymorphism with HNC risk (Fig. 2).
Similar results were observed in the subgroups regarding ethnicity, source of controls, sample size, tumor type and smoking status. Nevertheless, in the subgroup analysis concerning drinking status, elevated risk was shown in the heavy drinking subgroup under the homozygote comparison model (OR = 3.11; 95%CI = 1.52-3.02), indicating that TT carriers who have a heavy drinking history might have an increased HNC risk.
Sensitivity analysis and Bias diagnostics
To test the stability of the overall results, we first excluded the studies whose genetic distributions in controls markedly deviated from HWE, given that the deviation might result in any bias45. The significance of the overall data in the three models was also not statistically altered. Then, we also conducted one-way sensitivity analysis46 by deleting a single study each time. The statistical significance of the results was not also altered (data not shown), confirming the stability and credibility of the results.
Funnel plots were created for assessment of possible publication bias. Then, Egger’s linear regression tests were used to assess the symmetry of the plots. As a result, the data suggest that the funnel plots were symmetrical for the three models (homozygote comparison model: t = 0.13, P > 0.05; dominant model: t = −1.29, P > 0.05; recessive model: t = 0.87, P > 0.05), suggesting that the publication bias may have little influence on the results (Fig. 3).
Discussion
The effects of MTHFR C677T polymorphism on cancer risk were controversial. Recent published meta-analyses showed that 677 C > T variation might contribute to the development of breast and esophageal cancer47,48; however, a decreased cancer risk was observed in colorectal cancer49. Thus, MTHFR C677T polymorphism may play various roles in different carcinomas. In the present study, though the overall results failed to suggest a relationship between MTHFR C677T variation and HNC risk, the subgroup analysis indicated that homozygous 677TT might be associated with increased susceptibility to HNC in individuals who have a heavy drinking history.
Considering the possible effects of the confounding factors on the overall data, we tried to carry out subgroup analyses. In subgroup analysis according to ethnicity, no significant association between MTHFR 677T allele with HNC risk was observed among the three subgroups. The results were in line with the overall data, indicating that the effects of discrepancies among different ethnicities on the results were not evident.
When the data were stratified by source of controls and sample size, respectively, the results were not significantly different from the whole results, indicating that these two factors have little influence on the results. Likewise, in the subgroup stratified by tumor type, the results were in accordance with the overall data, implying that the dissimilarity of tumor types rarely affect the pooled results.
Whether chronic smoking interacts with folate status in the pathophysiologic process of disorders remains controversial50. Evidence suggests that low serum folic acid concentrations were commonly detected in smokers51. Nevertheless, cytological damage, an early event of carcinogenesis, is evident in the mouths of smokers, but it dose not correlate with folate status52. In the present meta-analysis, data about smoking status could be extracted from five included studies. The results showed that elevated risk was observed in neither the never-smoking subgroup nor the ever-smoking subgroup. The data failed to reveal an obvious interaction of smoking and MTHFR variation for HNC risk. However, when the data were divided by drinking status, elevated risk was shown in the heavy drinking subgroup under the homozygote comparison model. The data were in line with our previous meta-analysis about oral cancer11. The underlying mechanisms were unclear. MTHFR 677C → T polymorphism might play different roles for cancer risk according to the folate levels. It might reduce cancer risk when folate status is normal53 while lead to impaired stability and reduced activity of the enzyme under low folate conditions54. Notably, exposure to alcohol consumption might result in low folate status because chronic alcohol exposure may interfere with folate absorption by suppressing the folate carrier expression, thus decreasing the expression of folate transport proteins and reducing the hepatic uptake and renal conservation of circulating folate55,56. Also, alcohol might act as a folate antagonist that is responsible for abnormalities in folate-mediated one-carbon metabolism57. Moreover, folate depletion in mitochondria caused by chronic alcohol exposure might lead to abnormal DNA synthesis and DNA methylation that has been thought to be involved in the carcinogenesis process58,59. This might explain the interactions of alcohol with 677TT genotype in the genesis of HNC. However, only five of the selected studies concerned this issue. In the meantime, the data should be interpreted with care because of the limited sample sizes.
Evident heterogeneity was shown in every model for the overall data and thus, random-effect models were used to pool the data. Nevertheless, significant heterogeneities were removed in some subgroups while observed in other subgroups, indicating that the heterogeneities may be multifactorial. Besides the confounding factors considered in the subgroup analysis, other factors such as age, gender and life-style factors might contribute to the heterogeneities.
Several limitations might be involved in this meta-analysis. First, only publications written in English and Chinese were searched and selected. It is possible that articles written in other languages that might meet the inclusion criteria were missed. Thus, inevitable publication biases might exist though the funnel plots were tested to be symmetrical. Second, subgroup analyses regarding age, gender and other risk factors such as virus infection status and betel quid chewing have not been conducted because the data in the primary literature were insufficient. Third, hospital-based controls were used in some of the included studies, leading to non-differential misclassification bias. Moreover, the controls in some studies were not strictly matched to the cases. Therefore, any selection bias might have an influence on the overall results and future investigations with large sample sizes are required.
Despite the limitations, though the overall data of the present meta-analysis did not reveal an association of MTHFR C677T polymorphism with HNC risk, the subgroup analysis indicated that MTHFR 677TT alleles might increase HNC risk in individuals who have a heavy drinking history.
Additional Information
How to cite this article: Zhuo, X. et al. MTHFR C677T polymorphism interaction with heavy alcohol consumption increases head and neck carcinoma risk. Sci. Rep. 5, 10671; doi: 10.1038/srep10671 (2015).
References
Specenier, P. & Vermorken, J. B. Cetuximab: its unique place in head and neck cancer treatment. Biologics. 7, 77–90 (2013).
Liao, C. T. et al. Clinical evidence of field cancerization in patients with oral cavity cancer in a betel quid chewing area. Oral Oncol. 50, 721–731 (2014).
Machiels, J. P. et al. Advances in the management of squamous cell carcinoma of the head and neck. F1000Prime Rep. 6, 44 (2014).
James, S. J. et al. Mechanisms of DNA damage, DNA hypomethylation and tumor progression in the folate/methyl-deficient rat model of hepatocarcinogenesis. J. Nutr. 133, 3740S–3747S (2003).
Langie, S. A. et al. Maternal folate depletion and high-fat feeding from weaning affects DNA methylation and DNA repair in brain of adult offspring. FASEB J. 27, 3323–3334 (2013).
Trimmer, E. E. Methylenetetrahydrofolate reductase: biochemical characterization and medical significance. Curr Pharm Des. 19, 2574–2593 (2013).
Kim, Y. I. Role of the MTHFR polymorphisms in cancer risk modification and treatment. Future Oncol. 5, 523–542 (2009).
Toffoli, G. & De Mattia, E. Pharmacogenetic relevance of MTHFR polymorphisms. Pharmacogenomics 9, 1195–1206 (2008).
Rozen, R. Genetic predisposition to hyperhomocysteinemia: deficiency of methylenetetrahydrofolate reductase (MTHFR). Thromb Haemost. 78, 523–526 (1997).
Taioli, E. et al. Meta- and pooled analyses of the methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and colorectal cancer: a HuGE-GSEC review. Am J. Epidemiol. 170, 1207–1221 (2009).
Zhuo, X. et al. Polymorphisms of MTHFR C677T and A1298C association with oral carcinoma risk: a meta-analysis. Cancer Invest. 30, 447–452 (2012).
Higgins, J. P., Thompson, S. G., Deeks, J. J. & Altman, D. G. Measuring inconsistency in meta-analyses. BMJ. 327, 557–560 (2003).
Munafo, M. R., Clark, T. G. & Flint, J. Assessing publication bias in genetic association studies: evidence from a recent meta-analysis. Psychiatry Res. 129, 39–44 (2004).
Egger, M., Davey Smith, G., Schneider, M. & Minder, C. . Bias in meta-analysis detected by a simple, graphical test. BMJ. 315, 629–634 (1997).
Galbiatti, A. L. et al. Head and neck cancer: genetic polymorphisms and folate metabolism. Braz J Otorhinolaryngol. 78, 132–139 (2012).
Kane, M. A. The role of folates in squamous cell carcinoma of the head and neck. Cancer Detect Prev. 29, 46–53 (2005).
Zacho, J., Yazdanyar, S., Bojesen, S. E., Tybjaerg-Hansen, A. & Nordestgaard, B. G. Hyperhomocysteinemia, methylenetetrahydrofolate reductase c.677C>T polymorphism and risk of cancer: cross-sectional and prospective studies and meta-analyses of 75,000 cases and 93,000 controls. Int J Cancer 128, 644–652 (2011).
Dhawan, D., Panchal, H., Shukla, S. & Padh, H. Genetic variability & chemotoxicity of 5-fluorouracil & cisplatin in head & neck cancer patients: a preliminary study. Indian J. Med. Res. 137, 125–129 (2013).
Kawakita, D. et al. Association between dietary folate intake and clinical outcome in head and neck squamous cell carcinoma. Ann Oncol. 23, 186–192 (2012).
Zee, R. Y., Rose, L., Chasman, D. I. & Ridker, P. M. Genetic variation of fifteen folate metabolic pathway associated gene loci and the risk of incident head and neck carcinoma: the Women’s Genome Health Study. Clin. Chim. Acta. 418, 33–36 (2013).
Galbiatti, A. L. et al. Association between 11 genetic polymorphisms in folate-metabolising genes and head and neck cancer risk. Eur J. Cancer. 48, 1525–1531 (2012).
Cao, Y. et al. Polymorphisms of methylenetetrahydrofolate reductase are associated with a high risk of nasopharyngeal carcinoma in a smoking population from southern China. Mol. Carcinog. 49, 928–934 (2010).
Capaccio, P. et al. Association between methylenetetrahydrofolate reductase polymorphisms, alcohol intake and oropharyngolaryngeal carcinoma in northern Italy. J. Laryngol Otol. 119, 371–376 (2005).
Fard-Esfahani, P. et al. An increased risk of differentiated thyroid carcinoma in Iran with the 677C-->T homozygous polymorphism in the MTHFR Gene. Cancer Epidemiol. 35, 56–58 (2011).
Galbiatti, A. L. et al. Polymorphisms and haplotypes in methylenetetrahydrofolate reductase gene and head and neck squamous cell carcinoma risk. Mol. Biol. Rep. 39, 635–643 (2012).
Hsiung, D. T. et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 16, 108–114 (2007).
Kruszyna, L. et al. Polymorphic variants of folate metabolism genes and the risk of laryngeal cancer. Mol. Biol. Rep. 37, 241–247 (2010).
Kureshi, N., Ghaffar, S., Siddiqui, S., Salahuddin, I. & Frossard, P. M. Head and neck cancer susceptibility: a genetic marker in the methylenetetrahydrofolate reductase gene. ORL J. Otorhinolaryngol Relat Spec. 66, 241–245 (2004).
Neumann, A. S. et al. Methylenetetrahydrofolate reductase polymorphisms and risk of squamous cell carcinoma of the head and neck: a case-control analysis. Int. J. Cancer. 115, 131–136 (2005).
Ni, X. et al. Association between genetic polymorphisms in methylenetetrahydrofolate reductase and risk of laryngeal squamous cell carcinoma. Chin J. Otorhinolaryngol Head Neck Surg. 43, 435–438 (2008).
Ozdemir, S. et al. Increased T-allele frequency of 677 C>T polymorphism in the methylenetetrahydrofolate reductase gene in differentiated thyroid carcinoma. Genet Test Mol Biomarkers. 16, 780–784 (2012).
Prasad, V. V. & Wilkhoo, H. Association of the functional polymorphism C677T in the methylenetetrahydrofolate reductase gene with colorectal, thyroid, breast, ovarian and cervical cancers. Onkologie. 34, 422–426 (2011).
Reljic, A. et al. The methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and cancer risk: the Croatian case-control study. Clin Biochem. 40, 981–985 (2007).
Rodrigues, J. O. et al. Polymorphism of methylenetetrahydrofolate reductase (MTHFR) gene and risk of head and neck squamous cell carcinoma. Braz J. Otorhinolaryngol. 76, 776–782 (2010).
Sailasree, R., Nalinakumari, K. R., Sebastian, P. & Kannan, S. Influence of methylenetetrahydrofolate reductase polymorphisms in oral cancer patients. J. Oral. Pathol. Med. 40, 61–66 (2011).
Siraj, A. K. et al. Polymorphisms of selected xenobiotic genes contribute to the development of papillary thyroid cancer susceptibility in Middle Eastern population. BMC Med Genet. 9, 61 (2008).
Solomon, P. R., Selvam, G. S. & Shanmugam, G. Polymorphism in ADH and MTHFR genes in oral squamous cell carcinoma of Indians. Oral Dis. 14, 633–639 (2008).
Supic, G., Jovic, N., Kozomara, R., Zeljic, K. & Magic, Z. Interaction between the MTHFR C677T polymorphism and alcohol--impact on oral cancer risk and multiple DNA methylation of tumor-related genes. J. Dent. Res. 90, 65–70 (2011).
Suzuki, T. et al. One-carbon metabolism-related gene polymorphisms and risk of head and neck squamous cell carcinoma: case-control study. Cancer Sci. 98, 1439–1446 (2007).
Tsai, C. W. et al. Methylenetetrahydrofolate reductase (MTHFR) genotype, smoking habit, metastasis and oral cancer in Taiwan. Anticancer Res. 31, 2395–2399 (2011).
Vairaktaris, E. et al. Methylenetetrahydrofolate reductase polymorphism and minor increase of risk for oral cancer. J. Cancer Res. Clin. Oncol. 132, 219–222 (2006).
Vylliotis, A. et al. Effect of thrombosis-related gene polymorphisms upon oral cancer: a regression analysis. Anticancer Res. 33, 4033–4039 (2013).
Weinstein, S. J. et al. Folate intake, serum homocysteine and methylenetetrahydrofolate reductase (MTHFR) C677T genotype are not associated with oral cancer risk in Puerto Rico. J. Nutr. 132, 762–767 (2002).
Kweon, S. S., Shin, M. H., Kim, H. N., Kim, S. H. & Kang, H. C. Polymorphisms of methylenetetrahydrofolate reductase and glutathione S-transferase are not associated with the risk of papillary thyroid cancer in Korean population. Mol. Biol. Rep. 41, 3793–3799 (2014).
Thakkinstian, A., McElduff, P., D’Este, C., Duffy, D. & Attia, J. A method for meta-analysis of molecular association studies. Stat. Med. 24, 1291–1306 (2005).
Tobias, A. Assessing the influence of a single study in the meta-analysis estimate. Stata Techn. Bull. 8, 15–17 (1999).
Li, K., Li, W. & Dong, X. Association of 677 C>T (rs1801133) and 1298 A>C (rs1801131) polymorphisms in the MTHFR gene and breast cancer susceptibility: a meta-analysis based on 57 individual studies. PLoS One. 9, e71290 (e72014).
Wen, Y. Y., Yang, S. J., Zhang, J. X. & Chen, X. Y. Methylenetetrahydrofolate reductase genetic polymorphisms and esophageal squamous cell carcinoma susceptibility: a meta-analysis of case-control studies. Asian Pac. J. Cancer Prev. 14, 21–25 (2013).
Guo, X. P. et al. Association of MTHFR C677T polymorphisms and colorectal cancer risk in Asians: evidence of 12,255 subjects. Clin. Transl. Oncol. 16, 623–629 (2014).
Okumura, K. & Tsukamoto, H. Folate in smokers. Clin. Chim. Acta. 412, 521–526 (2011).
Tungtrongchitr, R. et al. Relationship of tobacco smoking with serum vitamin B12, folic acid and haematological indices in healthy adults. Public Health Nutr.. 6, 675–681 (2003).
Gabriel, H. E. et al. Chronic cigarette smoking is associated with diminished folate status, altered folate form distribution and increased genetic damage in the buccal mucosa of healthy adults. Am J. Clin. Nutr. 83, 835–841 (2006).
Ma, J. et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions and risk of colorectal cancer. Cancer Res. 57, 1098–1102 (1997).
Yamada, K., Chen, Z., Rozen, R. & Matthews, R. G. Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. Proc. Natl. Acad. Sci. USA. 98, 14853–14858 (2001).
Halsted, C. H., Villanueva, J. A., Devlin, A. M. & Chandler, C. J. Metabolic interactions of alcohol and folate. J. Nutr. 132, 2367S–2372S (2002).
Hutson, J. R., Stade, B., Lehotay, D. C., Collier, C. P. & Kapur, B. M. Folic acid transport to the human fetus is decreased in pregnancies with chronic alcohol exposure. PLoS One. 7, e38057 (2012).
Seitz, H. K., Maurer, B. & Stickel, F. Alcohol consumption and cancer of the gastrointestinal tract. Dig. Dis. 23, 297–303 (2005).
Biswas, A., Senthilkumar, S. R. & Said, H. M. Effect of chronic alcohol exposure on folate uptake by liver mitochondria. Am J. Physiol. Cell Physiol. 302, C203–C209 (2012).
Mason, J. B. & Choi, S. W. Effects of alcohol on folate metabolism: implications for carcinogenesis. Alcohol. 35, 235–241 (2005).
Author information
Authors and Affiliations
Contributions
X.Z. and Q.Z. planned and experiment designed; X.Z. and J.S. wrote the manuscript draft; D.L. and Y.W. processed data and prepared the figures and tables; All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhuo, X., Song, J., Li, D. et al. MTHFR C677T polymorphism interaction with heavy alcohol consumption increases head and neck carcinoma risk. Sci Rep 5, 10671 (2015). https://doi.org/10.1038/srep10671
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep10671
This article is cited by
-
Super-assembly of integrated gold magnetic assay with loop-mediated isothermal amplification for point-of-care testing
Nano Research (2023)
-
Interaction between alcohol consumption and methylenetetrahydrofolate reductase polymorphisms in thyroid cancer risk: National Cancer Center cohort in Korea
Scientific Reports (2018)
-
Alcohol and head and neck cancer
Cancer and Metastasis Reviews (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.