Abstract
Although most studies have reported that high serum lactate dehydrogenase (LDH) levels are associated with poor prognosis in several malignancies, the consistency and magnitude of the impact of LDH are unclear. We conducted the first comprehensive meta-analysis of the prognostic relevance of LDH in solid tumors. Overall survival (OS) was the primary outcome; progression-free survival (PFS) and disease-free survival (DFS) were secondary outcomes. We identified a total of 68 eligible studies that included 31,857 patients. High LDH was associated with a HR for OS of 1.48 (95% CI = 1.43 to 1.53; P < 0.00001; I2 = 93%), an effect observed in all disease subgroups, sites, stages and cutoff of LDH. HRs for PFS and DFS were 1.70 (95% CI = 1.44 to 2.01; P < 0.00001; I2 = 13%) and 1.86(95% CI = 1.15 to 3.01; P = 0.01; I2 = 88%), respectively. Analysis of LDH as a continuous variable showed poorer OS with increasing LDH (HR 2.11; 95% CI = 1.35 to 3.28). Sensitivity analyses showed there was no association between LDH cutoff and reported HR for OS. High LDH is associated with an adverse prognosis in many solid tumors and its additional prognostic and predictive value for clinical decision-making warrants further investigation.
Similar content being viewed by others
Introduction
Cancer is the leading cause of death in economically developed countries and the second leading cause of death in developing countries1. In the United States, a total of 1,660,290 new cancer cases and 580,350 cancer deaths were projected to occur in 20132. In Europe, there were an estimated 3.45 million new cases of cancer (excluding non-melanoma skin cancer) and 1.75 million deaths from cancer in 20123. Furthermore, the global burden of cancer continues to increase, largely because of population growth and increased life-expectancy3. Invasion and metastasis are two important hallmarks of cancer and are responsible for the majority of cancer deaths4. Although much effort has been devoted to the diagnosis and therapy of cancers, the overall prognosis is still unsatisfactory. A lack of knowledge of molecular biomarkers in cancer has limited the development of personalized therapies and improvements in survival. Therefore, there is an urgent need for universal, effective, readily available and inexpensive biomarkers in solid tumors to identify patients with a poor prognosis so that novel treatments can be initiated earlier.
The metabolism of cancer cells differs from that of normal cells. This is largely because cancer cells exhibit metabolic alterations that are frequently associated with reprogramming. Unlike normal cells, cancer cells preferentially metabolize glucose by glycolysis to generate sufficient energy for the demands of rapid proliferation, even in the presence of adequate oxygen5.This phenomenon is known as the Warburg effect and is one of the predominant metabolicalterations that occur during malignant transformation. In this process, transcriptional programs regulated by oncogenes stabilize hypoxia-inducible factor 1 alpha (HIF-1α). HIF-1α contributes to the upregulation of most enzymes involved in the glycolytic pathway, including lactate dehydrogenase (LDH).In the final step of aerobic glycolysis, LDH converts pyruvate tolactate, which is coupled with the oxidation of NADH to NAD+. These metabolic changes are reflected by an elevated serum LDH level6(hereinafter LDH).
Elevated LDH has been recognized as a poor prognostic indicator in cancer for many years7,8,9,10. LDH has also been incorporated in prognostic scores for several types of cancer11. However, the consistency and magnitude of the prognostic impact of LDH are unclear12,13,14. The aim of this study was to review published studies and use standard meta-analytic techniques to quantify the prognostic value of LDH in various solid tumors.
Methods
Data sources and searches
This analysis was conducted in line with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines15. PubMed was searched for studies evaluating the LDH and survival in solid tumors from 1978 to 2014. We used various medical subject heading terms, including “l-lactate dehydrogenase”, “prognosis”, “multivariate analysis” and “proportional hazard model”. Title/abstract words included “lactate dehydrogenase”, “LDH”, “prognosis”, “prognose”, “prognostic”, “multivariate analysis”, “proportional hazard model”, “COX proportional hazard model” and “COX models”. The full search strategy is described in the Supplementary Methods (available online).
Study selection
Inclusion criteria for the primary analysis were as follows: 1) studies of people with solid tumors reporting on the prognostic impact of LDH; 2) prospective or retrospective cohort design with a clearly defined source population and justifications for all excluded eligible cases; 3) sample size greater than 200; 4)statistical analysis using multivariate proportional hazards modeling that adjusted for clinical prognostic factors; and 5) reporting of the resultant adjusted hazard ratios (HRs) and their 95% confidence intervals (CIs) or a P value for overall survival (OS). For the secondary analyses, studies providing a HR for cancer-specific survival (CSS), progression-free survival (PFS), disease-free survival (DFS), or recurrence-free survival (RFS) were included as well.
Data extraction
OS was the primary outcome of interest. CSS, PFS and DFS were secondary outcomes. Two authors (J.Z. and H.W.) independently extracted information using predefined data abstraction forms. The following details were extracted: name of first author, year of publication, number of patients included in analysis, disease site, disease stage (non-metastatic, metastatic, mixed [both non-metastatic and metastatic]), study type (prospective or retrospective), cutoff defining high LDH and HRs and associated 95% confidence intervals for OS, PFS, DFS, or RFS as applicable. HRs were extracted preferentially from multivariate analyses where available. Where several HR values were given in an article, the value adjusted for most confounders was used.
Data synthesis
The meta-analysis was conducted initially for all included studies for each of the endpoints of interest. Subgroup analyses were conducted for predefined parameters such as disease site, disease stage and LDH cutoff and all data were limited to multivariate analyses. Disease site subgroups were generated if at least three studies on that site were available; the remaining studies were pooled in a subgroup termed “other.” LDH cutoff subgroups were < 250 U/L, 250–300 U/L, 301–400 U/L and >400 U/L. In three studies, the effect of LDH was reported as a continuous variable; we pooled those studies separately. Univariate meta-regression model analysis was performed to evaluate the relationship between covariates (LDH cutoff) and the HR for OS.
Statistical analyses
The meta-analysis was performed with RevMan 5.2 analysis software (Cochrane Collaboration, Copenhagen, Denmark). Estimates of HRs were weighted and pooled using the generic inverse-variance and random-effect model16. Analyses were conducted for all studies and differences between the subgroups were assessed using methods described by Deeks et al.17. Publication bias was assessed by visual inspection of the funnel plot. Heterogeneity was assessed using Cochran Q and I2 statistics. Meta-regression analysis was conducted using Stata12.0 software. All statistical tests were two-sided and statistical significance was defined as P less than 0.05. No correction was made for multiple testing.
Results
Description of studies
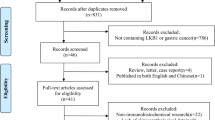
Sixty-eight studies were included in the meta-analysis. The selection process for the systematic review is shown in Figure S1 and the characteristics of the included studies are shown in Table 1. A total of 31,857 patients were included and the median trial sample size was 363.
Overall survival
Sixty-three studies comprising 29,620 patients reported HRs for OS. All studies analyzed LDH as a dichotomous variable. The studies have clearly shown that upper limit of normal (ULN) remains common for high LDH. The median cutoff for high LDH was 250U/L (range = 200–1000).
Two of the 63 eligible studies (3.2%) reported a non-statistically significant HR. A forest plot of all studies is presented in Figure 1. Overall, LDH greater than the cutoff was associated with a HR for OS of 1.48 (95% CI = 1.43 to 1.53; P < 0.00001). As the heterogeneity among studies was significant (P < 0.00001; I2 = 93%), a random-effects model was applied. To explore potential sources of heterogeneity, we performed subgroup analysis in the following subgroups: disease site, tumor stage and LDH subdivided by predefined cutoffs.
Forest plots showing HR for OS for LDH greater than or less than the cutoff.
HRs for each study are represented by the squares, the size of the square represents the weight of the study in the meta-analysis and the horizontal linecrossing the square represents the 95% confidenceinterval (CI). All statistical tests were two-sided.
The effect of LDH on OS among disease subgroups is shown in Figure 2. The prognostic effect of LDH was highest in renal cell carcinoma (HR = 1.84, 95% CI = 1.35 to 2.51), followed by nasopharyngeal carcinoma (HR = 1.82, 95% CI = 1.48 to 2.24), sarcoma (HR = 1.79, 95% CI = 1.30 to 2.47), melanoma (HR = 1.76, 95% CI = 1.56 to 1.98), prostate cancer (HR = 1.55, 95% CI = 1.06 to 2.26), colorectal cancer (HR = 1.52, 95% CI = 1.29 to 1.79) and lung cancer (HR = 1.50, 95% CI = 1.27 to 1.78). The HR for the subgroup of other unselected solid tumors was 1.69 (95% CI = 1.44 to 2.00). For the eight disease-site subgroups analyzed, there was statistically significant heterogeneity between disease sites (P < 0.00001), but no significant differences in the prognostic values of LDH between the subgroups (P for subgroup difference = 0.68).
The effect of LDH on OS among different disease stages is shown in Figure 3. The HRs were 1.54 (95% CI = 1.32 to 1.80) for non-metastatic disease, 1.70 (95% CI = 1.59 to 1.82) for metastatic disease and 1.20 (95% CI = 1.16 to 1.24) for a mixed group consisting of studies that included both metastatic and non-metastatic patients. There was statistically significant heterogeneity between disease stages (P < 0.00001). The prognostic value of LDH also varied significantly between different disease stages (P for subgroup difference < 0.00001).
The effect of LDH on OS among different cutoffs for LDH is shown in Figure 4. The HRs were 1.71 (95% CI = 1.38 to 2.12) for LDH cutoff < 250U/L, 1.67(95% CI = 1.52 to 1.84) for LDH cutoff 250 to 300U/L, 1.69 (95% CI = 1.27 to 2.24) for LDH cutoff 301 to 400U/L and 1.72(95% CI = 1.45 to 2.05)for LDH cutoff > 400 U/L. There was no statistically significant heterogeneity between the different cutoffs for LDH (P for subgroup difference = 0.99).
The scatter plot for the univariate meta-regression analysis is shown in Figure 5.A total of 63 studies was included in the meta-regression analysis. Overall, there was no statistically significant association between LDH cutoff and the HR for OS (P = 0.614).
Study-level (i.e., at the individual publication level) association of the cutoff used to define LDH and the HR for overall survival.
Each study is represented by a circle and the area of the circleis proportional to the number of patients enrolled in each study. The gradient of the dashed line represents the results of the meta-regression (β = 1.000138).
There was evidence of publication bias, with fewer small studies reporting negative results than would be expected (Supplementary Figure S2).
Three studies, comprising 1,766 patients, analyzed LDH as a continuous variable and reported HRs for OS. The pooled summary HR of these studies was 2.11 (95% CI, 1.35–3.28; P = 0.0003; I2 = 84%) per incremental LDH unit (Supplementary Figure S5).
Progression-free survival
Six studies, comprising 2,451 patients, reported HRs for PFS. Overall, LDH greater than the cutoff was associated with a HR for PFS of 1.70 (95% CI = 1.44 to 2.01; P < 0.00001; I2 = 13%). A forest plot is presented as Figure S3.
Disease-free (Recurrence-free) survival
A total of five trials, comprising 1,992 patients, reported HRs for DFS. Overall, LDH greater than the cutoff was associated with a HR for the endpoints of 1.86 (95% CI = 1.15 to 3.01; P = 0.01; I2 = 88%). A forest plot is presented in Figure S4.
Discussion
This is the first comprehensive meta-analysis of the prognostic relevance of LDH in solid tumors and it is based on a large pool of clinical studies (31,857 patients). We found a consistent effect of an elevated LDH on OS (HR = 1.48, 95%CI = 1.43 to 1.53) across all disease subgroups and stages. In addition, there is a trend toward a stronger prognostic value of LDH in metastatic disease compared with non-metastatic disease, which may reflect greater tumor burden. The prognostic impact of LDH on PFS and DFS (or RFS) is also robust. Interestingly, different cutoffs of LDH for different disease sites were reported in the included studies. However, the result of subgroups analysis for LDH cutoff showed that there was no association between LDH cutoff and reported HR for OS. This result was confirmed by meta-regression of LDH cutoff and HR for OS. Moreover, LDH was also related to poor prognosis in solid tumors when analyzed as a continuous variable. Our conclusions are supported by the fact that our selected studies were confined to those that used proportional hazards modeling to adjust for clinical prognostic factors and where the sample size was greater than 200.
There is a good biologic rationale for the use of LDH as a prognostic marker for cancer patients; however, the exact mechanism is not understood. One potential mechanism may be an association between LDH and the well-established phenomenon of oncogenicanaerobic glycolysis, or the Warburg effect5. This metabolic reprogramming is regulated by HIF-1α, as well as myc, through the transcriptional activation of key genes encoding metabolic enzymes; these include LDH, which converts pyruvate to lactate. This process is closely associated with an increased risk of invasion, metastasis and patient death77.
These analyses have several important implications. First, they show that a high LDH is associated with worse outcome, which suggests that LDH may be a useful biomarker to direct therapeutic selection78,79.This is because LDH is under the translational control of HIF-1α, as well as myc and thus is regulated by key oncogenic processes, such as the phosphatidylinositol 3-kinase/Akt/TORC1/hypoxia-inducible factor (PI3K/Akt/TORC1/HIF) pathway80,81,82. A recent study has demonstrated that the TORC1 inhibitor, temsirolimus, could provide therapeutic benefit in patients with RCC and high LDH79. Further work to investigate the predictive value of pretreatment LDH in other solid tumors may provide a more general insight into which patients derive benefit from TORC1 inhibition. Second, they show that increased LDH may be interpreted as reflecting high tumor burden or tumor aggressiveness. This suggests that dynamic changes of LDH level may be useful for predicting the prognosis in cancer patients after a primary operation, adjuvant chemotherapy, hormonal therapy, or radiotherapy65. Third, LDH allows the identification of a subgroup of tumors with a worse outcome. It is essential in the treatment of cancer to distinguish between low- and high-risk patients, thereby allowing stratification for standard or intensified treatment protocols. It has been shown that LDH can be used as an effective biomarker to guide the selection of regorafenib in patients with colorectal cancer; patients with high LDH may not be optimal candidates for regorafenib83.To adequately address these issues and dissect the complex relationship between LDH and cancer, future studies should be conducted within tumor- and stage-specific cohorts.
The strengths of this meta-analysis include the large sample size, estimation of HR using multivariate proportional hazards modeling that adjusted for clinical prognostic factors and analysis of a massive dataset comprising a large pool of clinical studies. LDH is also likely to be a cancer-specific biomarker, given that it is rarely increased in patients without cancer84. Thus, LDH may be a universal prognostic marker in cancer. To improve research in this area, studies with a more specific focus, such as those that address the impact of an individual LDH level on the prognosis of a homogeneous population of cancer patients (i.e., patients with the same cancer stage and subtype), would likely be more informative.
These analyses have limitations. One of the main limitations is the significant heterogeneity between studies, although we used random-effects models when pooling subgroup data. The heterogeneity in these studies could be explained by different patient characteristics or study designs. To facilitate interpretation, we grouped the patients by tumor type and tumor stage. Another limitation is that this is a literature-based analysis. It is compromised by the potential for publication bias, in which there is a tendency for predominantly positive results to have been published, thus inflating our estimate for the association between LDH and outcome. Our strict inclusion criteria (study size greater than 200, the requirement for HRs and a requirement for a 95% CI or P value) may have introduced selection bias. Most of the included studies were retrospective, which may have introduced reporting bias. Finally, different cutoffs used to assess high LDH level in these studies might also have contributed to the heterogeneity because it is possible that more false-positive cases were obtained with a cutoff of < 300 U/L than with a cutoff of >300 U/L. However, there is no accepted and validated absolute LDH level above which high LDH can be assigned. Instead, we used a cutoff of ULN. This may have introduced substantial heterogeneity, which may not have been fully accounted for by our use of sensitive analyses. The use of ULN is less robust; however, this was the only feasible method with the data available. An internationally accepted and validated LDH cutoff is warranted.
In summary, our data suggest that pretreatment LDH is a simple, cost-effective prognostic factor that can be considered as a criterion to consider patients in different prognostic groups. LDH is also a potential predictive marker to guide individual therapy decisions in solid tumors. Further, adequate, multi-center prospective studies are required to explore the clinical utility of LDH in solid tumors.
References
Mathers, C., Fat, D. M. & Boerma, J. T. The global burden of disease : 2004 update.1–146 (World Health Organization, Geneva, Switzerland; 2008).
Siegel, R., Naishadham, D. & Temal, A. Cancer statistics, 2013. CA Cancer J Clin 63, 11–30(2013).
Ferlay, J. et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer 49, 1374–1403 (2013).
Fidler, I. J. The pathogenesis of cancer metastasis: the ’seed and soil’ hypothesis revisited. Nat Rev Cancer 3, 453–458 (2003).
Hsu, P. P. & Sabatini, D. M. Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 (2008).
Serganova, I. et al. Metabolic imaging: a link between lactate dehydrogenase A, lactate and tumor phenotype. Clin Cancer Res 17, 6250–6261 (2011).
Motzer, R. J. et al. Prognostic factors for survival in 1059 patients treated with sunitinib for metastatic renal cell carcinoma. Br J Cancer 108, 2470–2477 (2013).
Wan, X. B. et al. High pretreatment serum lactate dehydrogenase level correlates with disease relapse and predicts an inferior outcome in locally advanced nasopharyngeal carcinoma. Eur J Cancer 49, 2356–2364 (2013).
Mekenkamp, L. J. et al. Mucinous adenocarcinomas: poor prognosis in metastatic colorectal cancer. Eur J Cancer 48, 501–509 (2012).
Giroux, L. E. et al. Factors associated with long-term survival of patients with advanced non-small cell lung cancer. Respirology 17, 134–142 (2012).
Lorch, A. et al. Prognostic factors in patients with metastatic germ cell tumors who experienced treatment failure with cisplatin-based first-line chemotherapy. J Clin Oncol 28, 4906–4911 (2010).
Kamiya, N. et al. Clinical outcomes by relative docetaxel dose and dose intensity as chemotherapy for Japanese patients with castration-resistant prostate cancer: a retrospective multi-institutional collaborative study. Int J Clin Oncol 19, 157–164 (2014).
He, W. Z. et al. Gamma-glutamyl transpeptidase level is a novel adverse prognostic indicator in human metastatic colorectal cancer. Colorectal Dis 15, e443–e452 (2013).
Sau, S., Biswas, A., Roy, A., Sau, S. & Ganguly, S. Retrospective analysis of the clinical and demographic variables on the outcomes after second-line treatment in advanced non-small cell lung cancer. Indian J Med Paediatr Oncol 34, 274–279 (2013).
Liberati, A. et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med 6, e1000100 (2009).
Cochrane Handbook for Systematic Reviews of Interventions. http://www.cochrane.org/training/cochrane-handbook Accessed October. 24, 2014 (2011).
Higgins, J. P. T. & Green, S. (ed.) Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (updated March 2011). (The Cochrane Collaboration., 2011).
Hannisdal, E., Fossa, S. D. & Host, H. Blood tests and prognosis in bladder carcinomas treated with definitive radiotherapy. Radiother Oncol 27, 117–122 (1993).
Kawahara, M. et al. Prognostic factors and prognostic staging system for small cell lung cancer. Jpn J Clin Oncol 27, 158–165 (1997).
Tonini, G. P. et al. MYCN oncogene amplification in neuroblastoma is associated with worse prognosis, except in stage 4s: the Italian experience with 295 children. J Clin Oncol 15, 85–93 (1997).
Eton, O. et al. Prognostic factors for survival of patients treated systemically for disseminated melanoma. J Clin Oncol 16, 1103–1111 (1998).
Tamura, M. et al. Prognostic factors of small-cell lung cancer in Okayama Lung Cancer Study Group Trials. Acta Med Okayama 52, 105–111 (1998).
Lagerwaard, F. J. et al. Identification of prognostic factors in patients with brain metastases: a review of 1292 patients. Int J Radiat Oncol Biol Phys 43, 795–803 (1999).
Motzer, R. J. et al. Survival and prognostic stratification of 670 patients with advanced renal cell carcinoma. J Clin Oncol 17, 2530–2540 (1999).
Scher, H. I. et al. Post-therapy serum prostate-specific antigen level and survival in patients with androgen-independent prostate cancer. J Natl Cancer Inst 91, 244–251 (1999).
Bacci, G. et al. Prognostic factors in nonmetastatic Ewing’s sarcoma of bone treated with adjuvant chemotherapy: analysis of 359 patients at the Istituto Ortopedico Rizzoli. J Clin Oncol 18, 4–11 (2000).
Vigano, A. et al. Clinical survival predictors in patients with advanced cancer. Arch Intern Med 160, 861–868 (2000).
Pierga, J. Y. et al. Effect of adjuvant chemotherapy on outcome in patients with metastatic breast carcinoma treated with first-line doxorubicin-containing chemotherapy. Cancer 91, 1079–1089 (2001).
Motzer, R. J., Bacik, J., Murphy, B. A., Russo, P. & Mazumdar, M. Interferon-alfa as a comparative treatment for clinical trials of new therapies against advanced renal cell carcinoma. J Clin Oncol 20, 289–296 (2002).
Atzpodien, J., Royston, P., Wandert, T. & Reitz, M. Metastatic renal carcinoma comprehensive prognostic system. Br J Cancer 88, 348–353 (2003).
Han, C. et al. Comparison of prognostic factors in patients in phase I trials of cytotoxic drugs vs new noncytotoxic agents. Br J Cancer 89, 1166–1171 (2003).
Polee, M. B. et al. Prognostic factors for survival in patients with advanced oesophageal cancer treated with cisplatin-based combination chemotherapy. Br J Cancer 89, 2045–2050 (2003).
Aoe, K. et al. Thrombocytosis as a useful prognostic indicator in patients with lung cancer. Respiration 71, 170–173 (2004).
Bacci, G. et al. Prognostic significance of serum lactate dehydrogenase in osteosarcoma of the extremity: experience at Rizzoli on 1421 patients treated over the last 30 years. Tumori 90, 478–484 (2004).
Aoe, K. et al. Serum hemoglobin level determined at the first presentation is a poor prognostic indicator in patients with lung cancer. Intern Med 44, 800–804 (2005).
Giaccone, G. et al. Phase III study of adjuvant vaccination with Bec2/bacille Calmette-Guerin in responding patients with limited-disease small-cell lung cancer (European Organisation for Research and Treatment of Cancer 08971–08971B; Silva Study). J Clin Oncol 23, 6854–6864 (2005).
Cook, R. J. et al. Markers of bone metabolism and survival in men with hormone-refractory metastatic prostate cancer. Clin Cancer Res 12, 3361–3367 (2006).
Bacci, G. et al. Ewing’s sarcoma family tumours. Differences in clinicopathological characteristics at presentation between localised and metastatic tumours. J Bone Joint Surg Br 89, 1229–1233 (2007).
Escudier, B. et al. Prognostic factors of metastatic renal cell carcinoma after failure of immunotherapy: new paradigm from a large phase III trial with shark cartilage extract AE 941. J Urol 178, 1901–1905 (2007).
Gripp, S. et al. Survival prediction in terminally ill cancer patients by clinical estimates, laboratory tests and self-rated anxiety and depression. J Clin Oncol 25, 3313–3320 (2007).
Laurie, S. A. et al. The impact of anemia on outcome of chemoradiation for limited small-cell lung cancer: a combined analysis of studies of the National Cancer Institute of Canada Clinical Trials Group. Ann Oncol 18, 1051–1055 (2007).
Saito, T., Hara, N., Kitamura, Y. & Komatsubara, S. Prostate-specific antigen/prostatic acid phosphatase ratio is significant prognostic factor in patients with stage IV prostate cancer. Urology 70, 702–705 (2007).
Schmidt, H. et al. Pretreatment levels of peripheral neutrophils and leukocytes as independent predictors of overall survival in patients with American Joint Committee on Cancer Stage IV Melanoma: results of the EORTC 18951 Biochemotherapy Trial. J Clin Oncol 25, 1562–1569 (2007).
Bedikian, A. Y. et al. Prognostic factors that determine the long-term survival of patients with unresectable metastatic melanoma. Cancer Invest 26, 624–633 (2008).
Neuman, H. B. et al. A single-institution validation of the AJCC staging system for stage IV melanoma. Ann Surg Oncol 15, 2034–2041 (2008).
Hashimoto, K. et al. Do recurrent and metastatic pancreatic cancer patients have the same outcomes with gemcitabine treatment? Oncology-Basel 77, 217–223 (2009).
Culp, S. H. et al. Can we better select patients with metastatic renal cell carcinoma for cytoreductive nephrectomy? Cancer 116, 3378–3388 (2010).
Kim, S. T. et al. Prognostic model to predict outcomes in non-small cell lung cancer patients with erlotinib as salvage treatment. Oncology-Basel 79, 78–84 (2010).
Suh, S. Y. et al. Construction of a new, objective prognostic score for terminally ill cancer patients: a multicenter study. Support Care Cancer 18, 151–157 (2010).
Tanrikulu, A. C. et al. A clinical, radiographic and laboratory evaluation of prognostic factors in 363 patients with malignant pleural mesothelioma. Respiration 80, 480–487 (2010).
Bedikian, A. Y. et al. Predictive factors for the development of brain metastasis in advanced unresectable metastatic melanoma. Am J Clin Oncol 34, 603–610 (2011).
Chibaudel, B. et al. Simplified prognostic model in patients with oxaliplatin-based or irinotecan-based first-line chemotherapy for metastatic colorectal cancer: a GERCOR study. Oncologist 16, 1228–1238 (2011).
Feliu, J. et al. Development and validation of a prognostic nomogram for terminally ill cancer patients. J Natl Cancer Inst 103, 1613–1620 (2011).
Sougioultzis, S. et al. Palliative gastrectomy and other factors affecting overall survival in stage IV gastric adenocarcinoma patients receiving chemotherapy: a retrospective analysis. Eur J Surg Oncol 37, 312–318 (2011).
Armstrong, A. J., George, D. J. & Halabi, S. Serum lactate dehydrogenase predicts for overall survival benefit in patients with metastatic renal cell carcinoma treated with inhibition of mammalian target of rapamycin. J Clin Oncol 30, 3402–3407 (2012).
Bidard, F. C. et al. Assessment of circulating tumor cells and serum markers for progression-free survival prediction in metastatic breast cancer: a prospective observational study. Breast Cancer Res 14, R29 (2012).
Jakob, J. A. et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer 118, 4014–4023 (2012).
Li, G. et al. Increased pretreatment levels of serum LDH and ALP as poor prognostic factors for nasopharyngeal carcinoma. Chin J Cancer 31, 197–206 (2012).
Shinohara, N. et al. A new prognostic classification for overall survival in Asian patients with previously untreated metastatic renal cell carcinoma. Cancer Sci 103, 1695–1700 (2012).
Weide, B. et al. Serum markers lactate dehydrogenase and S100B predict independently disease outcome in melanoma patients with distant metastasis. Br J Cancer 107, 422–428 (2012).
Zhou, G. Q. et al. Baseline serum lactate dehydrogenase levels for patients treated with intensity-modulated radiotherapy for nasopharyngeal carcinoma: a predictor of poor prognosis and subsequent liver metastasis. Int J Radiat Oncol Biol Phys 82, e359–e365 (2012).
Du, J. et al. High preoperative plasma fibrinogen is an independent predictor of distant metastasis and poor prognosis in renal cell carcinoma. Int J Clin Oncol 18, 517–523 (2013).
Durnali, A. et al. Prognostic factors for teenage and adult patients with high-grade osteosarcoma: an analysis of 240 patients. Med Oncol 30, 624 (2013).
Giessen, C. et al. Evaluation of prognostic factors in liver-limited metastatic colorectal cancer: a preplanned analysis of the FIRE-1 trial. Br J Cancer 109, 1428–1436 (2013).
Jin, Y. et al. Serum lactic dehydrogenase strongly predicts survival in metastatic nasopharyngeal carcinoma treated with palliative chemotherapy. Eur J Cancer 49, 1619–1626 (2013).
Powles, T., Bascoul-Mollevi, C., Kramar, A., Lorch, A. & Beyer, J. Prognostic impact of LDH levels in patients with relapsed/refractory seminoma. J Cancer Res Clin Oncol 139, 1311–1316 (2013).
Shinohara, N. et al. Is Memorial Sloan-Kettering Cancer Center risk classification appropriate for Japanese patients with metastatic renal cell carcinoma in the cytokine era? Urol Oncol 31, 1276–1282 (2013).
van Kessel, C. S. et al. Radiological heterogeneity in response to chemotherapy is associated with poor survival in patients with colorectal liver metastases. Eur J Cancer 49, 2486–2493 (2013).
Weide, B. et al. Serum S100B, lactate dehydrogenase and brain metastasis are prognostic factors in patients with distant melanoma metastasis and systemic therapy. PLoS One 8, e81624 (2013).
Halabi, S. et al. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol 32, 671–677 (2014).
Meckbach, D. et al. BRAF-V600 mutations have no prognostic impact in stage IV melanoma patients treated with monochemotherapy. PLoS One 9, e89218 (2014).
Poprach, A. et al. Clinical and laboratory prognostic factors in patients with metastatic renal cell carcinoma treated with sunitinib and sorafenib after progression on cytokines. Urol Oncol 32, 488–495 (2014).
Templeton, A. J. et al. Simple prognostic score for metastatic castration-resistant prostate cancer with incorporation of neutrophil-to-lymphocyte ratio. Cancer 120, 3346–3352 (2014).
Wang, X., Jiang, R. & Li, K. Prognostic significance of pretreatment laboratory parameters in combined small-cell lung cancer. Cell Biochem Biophys 69, 633–640 (2014).
Wei, Z., Zeng, X., Xu, J., Duan, X. & Xie, Y. Prognostic value of pretreatment serum levels of lactate dehydrogenase in nonmetastatic nasopharyngeal carcinoma: single-site analysis of 601 patients in a highly endemic area. Onco Targets Ther 7, 739–749 (2014).
Yamaguchi, T. et al. Multicenter retrospective analysis of systemic chemotherapy for advanced neuroendocrine carcinoma of the digestive system. Cancer Sci 105,1176–1181 (2014).
Halabi, S. et al. Prognostic model for predicting survival in men with hormone-refractory metastatic prostate cancer. J Clin Oncol 21,1232–1237 (2003).
Schellhammer, P.F. et al. Lower baseline prostate-specific antigen is associated with a greater overall survival benefit from sipuleucel-T in the Immunotherapy for Prostate Adenocarcinoma Treatment (IMPACT) trial. Urology 81,1297–1302 (2013).
D'Amico, A.V. Chen, M.H. Cox, M.C. Dahut, W. & Figg, W.D. Prostate-specific antigen response duration and risk of death for patients with hormone-refractory metastatic prostate cancer. Urology 66,571–576 (2005).
Vaupel, P. & Mayer, A. Hypoxia in tumors: pathogenesis-related classification, characterization of hypoxia subtypes and associated biological and clinical implications. Adv Exp Med Biol 812, 19–24 (2014).
Scartozzi, M. et al. Pre-treatment lactate dehydrogenase levels as predictor of efficacy of first-line bevacizumab-based therapy in metastatic colorectal cancer patients. Br J Cancer 106, 799–804 (2012).
Armstrong, A. J., George, D. J. & Halabi, S. Serum lactate dehydrogenase predicts for overall survival benefit in patients with metastatic renal cell carcinoma treated with inhibition of mammalian target of rapamycin. J Clin Oncol 30, 3402–3407 (2012).
Kim, J. W. & Dang, C. V. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci 30, 142–150 (2005).
Kim, J. W. & Dang, C. V. Cancer’s molecular sweet tooth and the Warburg effect. Cancer Res 66, 8927–8930 (2006).
Majumder, P. K. et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med 10, 594–601 (2004).
Del Prete, M. et al. LDH serum levels as a predictive factor for global outcome in pretreated colorectal cancer patients receiving regorafenib: Implications for clinical management. J Clin Oncol 32, 2318 (2014).
Harrison, D. E. et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395 (2009).
Acknowledgements
None
Author information
Authors and Affiliations
Contributions
Conception and design: J.Z. and H.W. Collection and checking eligible studies included in the meta-analysis: J.Z. and Y.Y. Acquisition of data: J.Z. and Y.Y. Analysis of data: J.Z., Y.Y., B.L., Q.Y., P.Z. and H.W. Statistical analyses: J.Z., Y.Y. and B.L. Writing of manuscript: J.Z. and H.W. Preparation of tables and figures: B.L., Q.Y. and P.Z. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, J., Yao, YH., Li, BG. et al. Prognostic value of pretreatment serum lactate dehydrogenase level in patients with solid tumors: a systematic review and meta-analysis. Sci Rep 5, 9800 (2015). https://doi.org/10.1038/srep09800
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep09800
This article is cited by
-
Analysis of clinical factors impacting recurrence in myxofibrosarcoma
Scientific Reports (2024)
-
Serum lactate dehydrogenase level predicts the prognosis in bladder cancer patients
BMC Urology (2023)
-
Prognostic value of serum α-HBDH levels in patients with lung cancer
World Journal of Surgical Oncology (2023)
-
Lactate dehydrogenase: relationship with the diagnostic GLIM criterion for cachexia in patients with advanced cancer
British Journal of Cancer (2023)
-
The key to immunotherapy: how to choose better therapeutic biomarkers for patients with non-small cell lung cancer
Biomarker Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.