Abstract
Studies have proven that IL-2 and IL-15 showed contrasting roles during CIK cells preparation. By employing microarray, we analyzed miRNA expression profiles of PBMC, CIKIL-2 and CIKIL-15. Advanced bioinformatic analyses were performed to explore the key miRNAs which may regulate cell proliferation and anti-tumor activity of CIK. We identified 261 differentially expressed miRNAs (DEMs) between PBMC and CIKIL-2 and 249 DEMs between PBMC and CIKIL-15. MiR-143-3p/miR-145-5p was miRNA cluster which may positively regulate cell proliferation. In contrast, miR-340-5p/miR-340-3p cluster may negatively regulate cell proliferation via induction apoptosis, which may cause decreased cell proliferation capacity of CIKIL-2. MiRNA-target interaction analysis indicated that 10 co-downregulated miRNAs may synergistically turn on the expression of a pool of tumor cytotoxic genes in CIK cells. The DEMs between CIKIL-2 and CIKIL-15 may contribute to enhanced tumor cytotoxic capacity of CIKIL-2. Importantly, we found that repressed miR-193a-5p may regulate the expressions of inhibitory receptor KLRD1. The results of the validation assay have shown that KLRD1 were upregulated in CIK cells. Our findings have provided new insights into mechanisms of CIK cells production and tumor cytotoxic function and shed light on their safety for clinical trial.
Similar content being viewed by others
Introduction
Amazing scientific advances have been translated into better ways to prevent, detect, diagnose and treat cancer during the past five years1. Nowadays, people are surviving longer after their cancer has been diagnosed due to these remarkable progress. Numerous therapeutics against cancer have shown large potential in clinical trials1. Notably, one group of strategies against cancer which are likely to revolutionize the treatment of certain cancer in the very near future are immunotherapies1. These therapeutics educate the patients' immune system to attack their cancer cells yielding both strong and durable response. Among these strategies, adoptive immunotherapy has shown great promise and encouraging efficacy in the tumor treatment with minimal adverse events2,3. Cytokine-induced killer (CIK) cells based immunotherapy is widely performed for clinical trials in China which is alternatives to conventional therapies2. CIK cells, a subset of T lymphocytes with a natural killer T cell phenotype, have been proven to be effective to most of tumors in vitro and in vivo4. CIK cells are generated from peripheral blood lymphocytes through time sequential stimulations of IFN-γ, monoclonal antibody against CD3 (OKT3) and IL-2. During this time period of CIK cells preparation, OKT3 provided mitogenic signals to T lymphocytes5. Priming with IFN-γ is to activate the monocytes through providing contact-dependent (CD58/LFA-3) and soluble (IL-12) crucial signals to promote generation of autophagy and antigen cross-presentation6. IL-2 is essential for T cell proliferation, survival and acquisition of cytolytic capacity in the following culture. At the end of expansion, a heterogeneous population of CD3+CD56+ CIK cells presenting potent cytotoxicity against a variety of tumor cells were obtained. However, the protocol for preparation of CIK cells can be differed for the purpose of enhancing the tumor cytotoxicity and CIK cells proliferation capacity7. It has been reported that the addition of IL-6 every 2–3 days during the preparation of CIK cells could inhibit the generation of Foxp3+ Treg cells and increase the proportion of CD3+CD56+ cells8. In our previous study, we have shown that CIK cells stimulated with combination of IL-2 and IL-15 exhibited enhanced proliferation capacity and cytotoxicity against lung cancer9. Interestingly, the results have indicated that CIK cells induced with combination of IL-2 and IL-15 could upregulate the expression levels of IFN-γ and TNF-α in mice models. In further investigation, we have found that CIKIL-2 showed greater tumor cytotoxicity than CIKIL-15 and CIKIL-15 exhibited enhanced proliferation capacity than CIKIL-210. By advanced bioinformatic analysis of RNA-seq data from CIKIL-2 and CIKIL-15, results indicated that genes participating in Wnt signal pathway and focal adhesion were upregulated in CIKIL-15 and the expression levels of genes involved in cytokine-cytokine receptor interaction were increased in CIKIL-210. Although the expression profiles of important genes in CIKIL-2 and CIKIL-15 have been well revealed, the regulation of these genes by IL-2 and IL-15 are still unknown.
MicroRNAs (miRNAs), a class of highly conserved ~20–22 nt long noncoding RNA, are essential molecules of post-transcriptional regulation of gene expression11. MiRNAs regulate gene expression negatively by targeting the 3′ untranslated region (3'UTR) or coding region of the mRNA, leading to either RNA degradation or inhibition of translation12. MiRNAs participated in many biological processes including cell proliferation, differentiation, apoptosis and tumorgenesis13. More recently, it was reported that miRNAs are involved in regulatory networks in immune system and regulation of development of immune cells14. However, the regulatory functions of miRNAs in CIK cells expansion and acquisition of cytotoxic capacity have not been reported yet.
In order to identify the roles of miRNAs in regulatory network of CIK cells generation, we performed miRNAs microarray analysis between PBMC (peripheral blood mononuclear cell) and CD3+CD56+ CIK cells and investigated the changes in global miRNAs expression level. Advanced system biology strategies have been employed to comprehensively investigate the molecular mechanism of translational modulation of miRNAs during CIK cells expansion. Our finding will provide evidence to better understand the acquisition of tumor cytotoxicity and proliferation capacity of CIK cells.
Results
Dynamic miRNA profiles between PBMC and CIK cells
We have prepared CIKIL-2 and CIKIL-15 from PBMCs of three healthy volunteers under identical conditions. Sequentially, PBMCs, CIKIL-2 and CIKIL-15 were sampled and preserved in liquid nitrogen for the following miRNA microarray analysis. The phenotype of CIK cells were determined by flow cytometry. The result showed that the average percentages of CD3+CD56+ cells were over 98% in both CIKIL-2 and CIKIL-15 (Figure S1 and Table S1). After confirmation of the portion of CD3+CD56+ cells, the proliferation capacity and tumor cytotoxicity of both CIKIL-2 and CIKIL-15 were measured by automatic absolute cell counting and CCK-8 based method respectively. The results of these two assays were previously described10. Then, miRNAs was isolated and purified from the preserved PBMC, CIKIL-2 and CIKIL-15. After quality assessment, the miRNAs were labeled and hybridized to Agilent V19.0 miRNA array. The array contained 2,006 human miRNAs, which allowed us to perform a deep investigation to CIK cells miRNA expression. After normalization of the raw data, we screened 261 differentially expressed miRNAs (DEMs) between PBMC and CIKIL-2 and 249 DEMs between PBMC and CIKIL-15 by the following criteria: fold change (FC) > 2 or FC < 0.5, P value < 0.05, all flag signals of 3 replicate were all same (Table S2 and S3). Of these DEMs between PBMC and CIKIL-2, 111 and 150 miRNAs were downregulated and upregulated respectively (Figure 1A). There were 109 downregulated and 140 upregulated miRNAs between PBMC and CIKIL-15 (Figure 1A). However, no significant DEM was identified between CIKIL-2 and CIKIL-15 by the screening criteria referred above. By further comparison of the miRNA expression patterns of CIKIL-2 and CIKIL-15 against PBMC, we found that 130 miRNAs were co-upregulated, accounting for 86.66% and 92.85% of total upregulated miRNAs in CIKIL-2 and CIKIL-15 respectively. In addition, there were 103 miRNAs co-downregulating in both of the two CIK cells, accounting for 92.79% and 94.49% of total downregulated miRNAs in CIKIL-2 and CIKIL-15 respectively (Figure 1B).
Differentially expressed miRNA chromosome clustering
Interestingly, evidences have supported that CIKIL-2 and CIKIL-15 shared over 90% of downregulated miRNAs. Furthermore, target genes may be upregulated in response to downregulation of corresponding miRNA. Therefore, co-downregulated miRNAs were chosen for further analysis. In order to identify their function during CIK preparation, we aligned the co-downregulated miRNAs to the chromosomes they located based on the chromosome coordinates of each miRNA (Figure 2A). We assumed that miRNAs which closed to each other may have the same biological function15,16. Among co-downregulated miRNAs, we have found 17 miRNA clusters in which the distance of miRNAs are no longer than 5000 bp (Figure 2A, Table 1). The heatmap and hierarchical analysis of 17 DEMs clusters were shown in Figure 2B. There were 3 miRNA clusters in co-upregulated miRNAs. No miRNA cluster were observed in CIKIL-2 or CIKIL-15 specific DEMs.
Chromosome clustering and gene ontology analysis (A) Chromosome clustering analysis of co-downregulated miRNAs. Red lines and green lines represent co-downregulated miRNAs and background miRNAs on each chromosome respectively; (B) Heatmap and hierarchical clustering of differentially expressed miRNAs which are chosen for chromosome clustering and miRNA-targets interaction network analysis. (C) Significant gene ontology analysis of 3 co-downregulated miRNAs clusters including miR-29b-3p/miR-29c-3p, miR-143-3p/miR-145-5p and miR-340-5p/miR-340-3p. P value < 0.05 for all significant GO terms. (D) 2-D view analyses establish associations between GO terms participating in promoting cell proliferation and genes regulated by miR-143-3p/miR-145-5p cluster. (E) 2-D view analyses between induction of apoptosis (GO term) and genes regulated by miR-340-5p/miR-340-3p.
Predicted target gene ontology and pathway analysis
To further characterize the function of the miRNA we identified, the target genes of each miRNA cluster were predicted based on miRTarBase Release 4.5. The target genes of each miRNA in miRTarBase were validated by Reporter assay, Western Blot, Microarray or pSILAC. Next, we performed gene ontological analysis to screen co-downregulated miRNA clusters whose target genes may be involved in cell proliferation and tumor cytotoxicity of CIK cells by employing annotation tool of DAVID bioinformation database. By analyzing the significant GO terms, we found that response to cytokine stimulus and positive regulation of cell proliferation were the most significant GO terms of the target genes regulated by miR-29b-3p/miR-29c-3p (C2) and miR-143-3p/miR-145-5p (C3) respectively (Figure 2C). Importantly, induction of apoptosis was significant among target genes of miR-340-5p/miR-340-3p (C4) (Figure 2C). We focused on cell proliferation and induction of apoptosis which are two key biological functions of CIK cells. Functional classification analysis in 2-D view was performed in order to visualize the associations between target genes of C3 and C4 clusters and GO terms including cell proliferation and induction of apoptosis (Figure 2D, Figure 2E). With respect to miR-143-3p/miR-145-5p cluster, HRAS, KRAS and NRAS which are the most common members of Ras subfamily were found to be strongly correlated with cell proliferation of CIK cells. By 2-D view analysis, two proto-oncogenes including Bcl2 and c-Myc which are involved in promoting cell proliferation were identified to be regulated by miR-143-3p/miR-145-5p cluster (Figure 2D). Among the genes regulated by miR-340-5p/miR-340-3p, TNFRSF10B was shown to participate in cell death (Figure 2E). To further explore the influence of co-downregulated miRNA cluster on function of CIK cells, we performed pathway analysis based on KEGG and Biocarta databases by using Fisher exact test. The results indicated that target genes of miR-29b-3p/miR-29c-3p, miR-143-3p/miR-145-5p and miR-23b-3p/miR-27b-3p clusters were all involved in IL-2 receptor beta chain in T cell activation and focal adhesion. The two pathways were significant in the target genes of each miRNA cluster (Figure 3A). By 2-D view analysis, the genes regulated by the 3 miRNA clusters were mainly involved in cell proliferation signaling transduction pulsed by IL-2 including kinases (JAK1, RAF1, CRKL and AKT1), signaling transducers (SOS1, HRAS, SOCS1, SOCS3 and IRS1), DNA binding proteins (FOS and E2F1) and apoptosis suppressors (BCL2 and BCL2L1) (Figure 3B). Among genes involved in focal adhesion, we found that components of extracellular matrix and their receptors were regulated by miR-29b-3p/miR-29c-3p including collagen, laminin and integrin (Figure 3C). Additionally, target genes regulated by miR-143-3p/miR-145-5p and miR-23b-3p/miR-27b-3p clusters were responsible for signal transduction which may be mediated by integrin.
Pathway analysis of co-downregulated miRNA clusters based on KEGG and Biocarta (A) Significant pathway analysis of miR-29b-3p/miR-29c-5p, miR-143-3p/miR-145-5p and miR-23b-3p/miR-27b-3p. P value < 0.05 for all significant pathways.; (B) 2-D view analysis of IL-2 receptor beta chain in T cell activation pathway. Associations between genes involved in this pathway and their regulatory miRNA clusters were shown; (C) 2-D view analysis of focal adhesion pathway. Green panes represent associations have already reported. Black panes represent associations have not been reported yet.
MiRNA-Target Interaction (MTI) network in tumor cytotoxicity pathway
Alternatively, we have employed miRTar database to identify miRNA-target interactions in cytotoxicity pathway. We inputted co-downregulated miRNAs and selected natural killer cell mediated cytotoxicity and cytokine-cytokine receptor interaction as our target pathways which were derived from KEGG database. We built the miRNA-target interaction network based on the anti-tumor factors expressed on cytotoxic lymphocytes (Figure 4A). The network analysis indicated that 10 distinct co-downregulated miRNA were found targeting cytotoxic genes and the hierarchical analysis of their expression profiles was shown in figure 2B. The results indicated that let-7c was a key miRNA which regulated 3 tumor toxic molecules including Fas ligand (FasL), TNFSF10 (Trail) and OSM (Oncostatin M). Among the cytotoxic genes, PRF1, GZMB, TNFSF10 and FasL were shown to be targeted only by co-downregulated miRNAs. Other anti-tumor genes including OSM, TNF-α and CD40LG were regulated by miRNAs of both downregulated and upregulated. In addition, we performed miRNAs-pathway interaction analysis to establish the linkage between these 10 miRNA and gene pool participating in natural killer cell mediated cytotoxicity pathway. By building the miRNA-pathway network, the results showed that 8 co-downregulated miRNAs were involved in regulation of natural killer cell mediated cytotoxicity pathway and the correlations between them were shown in figure 4B. In the gene group regulated by these 8 miRNAs, we found that receptors, signal transduction components and tumor cytotoxicity factors were all included (Figure 4C). Among receptors expressed on NK cells, inhibitory receptors (KIR2DL4; CD94) and activating receptors (ITGB2; KIR2DS5; NKG2C/E; NKp46) were identified which may work as sensors to discriminate normal cells and tumor cells.
MiRNA-target interactions network analysis of tumor cytotoxicity
(A) Regulatory network of tumor cytotoxic genes and shared differentially expressed miRNAs; (B) Interaction network between tumor cytotoxicity promoting miRNAs and components of natural killer cells mediated cytotoxicity pathway; (C) Potential impact of tumor cytotoxicity promoting miRNAs in natural killer cells mediated cytotoxicity pathway. Components of signaling pathway which were regulated by miRNAs we identified were marked in red and corresponding regulatory miRNAs were listed beside.
Validation of representative miRNAs and mRNAs
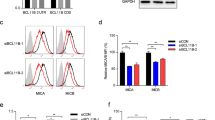
Next, we examined the expression profiles of co-downregulated miRNAs referred above by employing qRT-PCR across these 3 types of cells. Consistent with miRNA array data, the results have shown that all miRNAs we referred above were significantly downregulated in both CIKIL-2 and CIKIL-15 compared to PBMC (Figure 5A). Interestingly, among these 16 miRNAs, 10 miRNAs differentially expressed between CIKIL-2 and CIKIL-15, which have not been identified by microarray analysis (Figure 5A). Moreover, we validated the expression levels of cytotoxic genes in CIK cells in order to demonstrate the regulatory relationship between miRNAs of interest and their targets we suggested above. Except for OSM, the expression of all anti-tumor genes including TNF-α, PRF1, GZMB, FasL, TNFSF10 and CD40LG were significantly upregulated in CIK cells compared to PBMC, which were negatively correlated with expression profiles of their potential regulators (Figure 5B, Figure 6A and Figure 6B). Among these genes, the expression of GZMB and TNFSF10 were significantly higher in CIKIL-2 than CIKIL-15 in protein level (Figure 6A and Figure 6B). To further explore the cell proliferation mechanism of CIK cells, the expression of important genes regulated by miR-143-3p/miR-145-5p cluster were profiled. The results indicated that the expressions of c-Myc and Bcl-2 were significantly increased in CIKIL-2 and CIKIL-15 (Figure 5C and Figure 6B). Although NRAS mRNA was found to be significantly upregulated in CIK cells, no significant difference was observed in protein level (Figure 5C and Figure 6B). Importantly, we have examined the expression of inhibitory and activating receptors to evaluate the recognition specificity of CIK cells. NKG2D, which has been reported to be upregulated in CIK cells was taken as positive control in validation assay. By validation of both mRNA and protein level, the results showed only KLRD1 (CD94) were significantly upregulated in CIK cells (Figure 5D and Figure 6A).
qRT-PCR validation of relative expression levels of representative co-downregulated miRNAs and their targets.
(Data are plotted as mean ± SEM *P < 0.05; **P < 0.01; ***P < 0.0001; NS no significance) (A) Confirmation of the differentially expressed miRNAs by real-time PCR analysis. The miRNA expression was normalized with U6. The scale of Y-axis was presented in Log2Scale; (B) Validation of differentially expressed tumor cytotoxic genes between PBMC and CIK cells; (C) qRT-PCR analysis of the expression profiles of proto-oncogenes participating in cell proliferation of CIK cells; (D) Confirmation of differentially expressed inhibitory and activating receptors in PBMC, CIKIL-2 and CIKIL-15. NKG2D, which has been reported to be upregulated in CIK cells was taken as positive control. The expressions of protein coding genes were normalized with β-actin.
Flow cytometry and Western blot analysis of target genes.
(A) Flow cytometry analysis of the cytotoxic genes, inhibitory and activating receptors which are expressed on cell surface. Results were obtained in three independent samples. (B) PBMCs, CIKIL-2 and CIKIL-15 were lysed followed by cell total protein determination. The total protein amount were normalized for western blot analysis. Three independent samples were included.
Discussion
Nowadays, immunotherapies which have made remarkable progress are revolutionizing the treatment of cancers. However, immunotherapy development is a challenging work which need large amount of studies to prove its effectiveness and safety. CIK is one of adoptive immunotherapy approaches which have exhibited potent cytolytic activities against tumor cells with minimal adverse effects. The original work of CIK study was reported by Schmidt-Wolf from Stanford4. Clinical trials of CIK based immunotherapies were widely performed in China, however, few studies were observed which focused on molecular mechanism of tumor toxic function. Herein, we performed microarray analysis to identify the miRNA expression profiles of CD3+CD56+CIK cells for the first time and elucidate their proliferation and cytolytic mechanisms on post-transcriptional regulation level. The efficiency of CIK based immunotherapies were determined by cell proliferation and cytotoxicity capacities against tumor5. To improve the effectiveness of CIK cells, cytokines including IL-1, IL-17, IL-12 and IL-15 were used instead of IL-2 or in combination with IL-27. IL-2 and IL-15 have similar biological function in vitro and they shared receptor signaling components (IL-2/15Rβγc) with each other17,18. By pathway analysis, we found co-downregulated miRNA clusters including miR-29b-3p/miR-29c-3p, miR-143-3p/miR-145-5p and miR-23b-3p/miR-27b-3p were participated in IL-2 receptor beta chain in T cell activation (Figure 3A and 3B). However, contrasting roles of IL-2 and IL-15 were observed in lymphocytes based immunotherapy19. It was reported that CIK cells which were generated in combination with IL-2 and IL-15 showed greater cytotoxicity against lung cancer than CIK cells prepared with IL-2 alone9. Moreover, our previous comparative analysis indicated that CIKIL-15 showed enhanced cell proliferation capacity than CIKIL-2, whereas, CIKIL-2 showed greater tumor cytotoxic activity against tumor than CIKIL-15 in vivo10. Interestingly, advanced bioinformatic analysis and validation assays provided evidences to illuminate the potential mechanism of differential cell proliferation capacity and anti-tumor activity of CIKIL-2 and CIKIL-15.
We have identified 130 co-downregulated and 103 co-upregulated miRNAs through microarray analysis. By chromosome clustering and miRNA-target interaction analysis, we have focused on 16 miRNAs for further qRT-PCR validation assay. We have noticed that it was important to determine the abundance of miRNAs of interested because that very minimally expressed miRNA which showed dramatically change may be unimportant during biological process. Studies based on two distinct next-generation platforms have suggested that approximately 60 mature miRNA sequences comprised over 97% of miRNA sequences in mouse natural killer cells (NK cells)20,21. The results have shown that miR-21 was ranked as number 1 miRNA which accounted for nearly 31% of the miRNA sequence in NK cells21. Surprisingly, we have found that miR-29b, miR-27b, let-7c and miR-28 we included in this study were listed within the top 60 miRNAs which accounted for over 97% miRNA sequence in NK cells.
By chromosome clustering and GO analysis, we found that miR-143-3p/miR-145-5p were a miRNA cluster which may positively regulated the cell proliferation of CIK cells. Importantly, proto-oncogene including Ras family, Bcl-2 and c-Myc were predicted targets of this miRNA cluster. However, we found that only Bcl-2 and c-Myc were significantly upregulated during CIK generation (Figure 5C and Figure 6B). These two anti-apoptotic genes may be involved in cell proliferation during CIK preparation22. On the other hand, target genes of miRNA cluster containing miR-340-5p and miR-340-3p was identified to potentially participate in induction of apoptosis (Figure 2C and 2E). The results of qRT-PCR showed that the expression level of miR-340-5p and miR-340-3p were significantly higher in CIKIL-15 than CIKIL-2, which indicated that a pool of apoptosis promoting genes may be upregulated in CIKIL-2 compared to CIKIL-15. Furthermore, IL-15 is an anti-apoptotic cytokine which inhibits IL-2 mediated activation-induced cell death (AICD) of T cell and stimulates survival of memory T cell, whereas, IL-2 induces AICD and eliminates self-reactive T cell to maintain peripheral tolerance through upregulating of FasL and TNFSF1019,23. Interestingly, we have found that the expression of FasL and TNFSF10 were upregulated in both CIKIL-2 and CIKIL-15. However, the expression of TNFSF10 was higher in CIKIL-2 than CIKIL-15 (Figure 6B). Collectively, these evidences may explain our observation that CIKIL-15 showed greater proliferation potential than CIKIL-2 as previously described.
We have built the regulatory network among tumor toxic genes and differentially expressed miRNA which governed their expression. It showed that GZMB, which is a key anti-tumor molecule in vivo, was regulated mainly by miR-199a-5p and miR-199b-5p. Data from qRT-PCR and Western blot suggested that the expression level of GZMB was significantly increased in CIK cells. Importantly the results have implicated that the expression of GZMB was upregulated in CIKIL-2 compared to CIKIL-15, which negatively correlated with the expression profiles of miR-199a-5p and miR199b-5p and suggested the enhance tumor cytotoxicity of CIKIL-2. PRF1, which is known as pore-forming protein and induces apoptosis in synergy with GZMB was upregulated in both CIKIL-2 and CIKIL-1524,25. Except for TNF-α and TNFSF10 which have been reported as anti-tumor molecules of CIK, we have identified CD40LG as tumor toxic effector in CIK cells by analyzing interactions between co-downregulated miRNAs and cytokine-cytokine receptor interaction pathway26,27,28. The expression of CD40 LG was upregulated in CIK cells. Compared to PBMC, OSM was significantly downregulated in both CIKIL-2 and CIKIL-15. Studies have reported that OSM which was initially found to inhibit proliferation of several tumors appeared to promote growth of malignant cells now29,30,31. Therefore, evidences obtained from miRNA-Target network and validation assays have suggested the potential cytotoxicity mechanism of CIK cells. The higher expression of TNFSF10 and GZMB in CIKIL-2 may account for the greater tumor cytotoxic efficiency of CIKIL-2 than CIKIL-15.
Additionally, the function of natural killer cell is determined by the balance between signals triggered by activating and inhibitory receptors. CD94-NKG2A are important inhibitory receptor systems in most species and they transduce inhibitory signal through SHP-1 and SHP-232,33,34. By co-downregulated miRNAs-receptor interaction analysis, we found that KLRD1 (CD94) were significantly upregulated in CIK cells. KLRD1 is a peptide-selective receptor on NK cell, which binds HLA-E-peptide complex and provides inhibitory signal in absence of its signaling partner NKG2A35. This results suggested that increased expression of CD94 in CIK cell may protect HLA-E positive cell from lysis36.
In conclusion, we have performed microarray analysis to investigate the dynamic miRNA expression profiles during CIK cell preparation for the first time. By advanced bioinformatic analysis, the results indicated that miR-143-3p/miR-145-5p cluster may positively regulated cell proliferation through upregulating a group of proto-oncogenes. In contrast, miR-340-5p/miR-340-3p cluster may negatively regulated cell proliferation via induction apoptosis, which may cause decreased cell proliferation potential of CIKIL-2. MiRNA-target interaction analysis revealed that 10 co-downregulated miRNAs may synergistically promote the expression of a pool of tumor cytotoxic genes in CIK cells. Importantly, upregulation of inhibitory receptors (KLRD1) in CIK cells implicated the possibility that activation of CIK cells through activating receptors (NKG2D) could be negatively regulated by KLRD1. These evidences suggested that the presences of both activating and inhibitory receptors on CIK cells may contribute to their safety for clinical trial.
Methods
Antibodies and Cytokines
The antibodies for CIK cells phenotype assay were purchased from BD Biosciences. The antibodies used for detecting cell surface receptors and cytotoxic factors were obtained from BioLegend, Inc. and R&D System. For Western blot, antibodies were purchased from EMD Millipore and Santa Cruz Biotechnology, Inc. Cytokines for CIK cells preparation including OKT3, IFN-γ, IL-2 and IL-15 were from Miltenyi Biotec. Methods involving human peripheral blood in this studies were reviewed and approved by Bioethics Committee of Yan'an Affiliated Hospital of Kunming Medical University. The methods were carried out in accordance with the approved guidelines. Written informed consents have been given from all volunteers participated in this study.
Generation of CIK cells
The Bioethics Committee of Yan'an Affiliated Hospital of Kunming Medical University has approved the investigation protocols to draw blood from healthy volunteers after written informed consent for the purposes of CIK cells preparation against tumor and microarray analysis. The standard protocol of CIK generation was described previously9. Briefly, PBMCs were isolated by standard Ficoll separation and cultured in RPMI 1640 growth medium at a density of 5 × 106 cells/mL. The RPMI 1640 growth medium for CIK cell contained 10% FBS, 2% L-glutamine and antibiotics. The generation of CIK cells was primed by adding 1000 U/mL IFN-γ on day 0 and 100 ng/mL anti-CD3 antibody and 500 U/mL IL-2 or 10 ng/mL IL-15 within the following 15 days of culture. The CIK cells were propagated every 5 days with RPMI 1640 growth medium supplemented with anti-CD3 antibody and IL-2 or IL-15 respectively. The CIK cells were expanded for 15 days.
RNA isolation and labeling
Total RNA was extracted and purified using mirVana™ miRNA Isolation Kit (AM1560, Ambion, Austin, TX, US) following the manufacturer's instructions. The quality was assessed by an Agilent Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, US). MiRNA in total RNA was labeled by miRNA Complete Labeling and Hyb Kit (5190-0456, Agilent technologies, Santa Clara, CA, US) following the manufacturer's instructions.
Microarray hybridization
Labeled RNA samples were further detected by the miRNA array by using Agilent's human miRNA microarray, version 19.0. Each slide was hybridized with 100 ng Cy3-labeled RNA using miRNA Complete Labeling and Hyb Kit in hybridization Oven (G2545A, Agilent technologies, Santa Clara, CA, US) at 55°C, 20 rpm for 20 hours according to the manufacturer's instructions. After hybridization, slides were washed in staining dishes (121, Thermo Shandon, Waltham, MA, US) with Gene Expression Wash Buffer Kit (5188–5327, Agilent technologies, Santa Clara, CA, US).
Data acquisition and identification of differentially expressed miRNAs
Slides were scanned by Agilent Microarray Scanner (G2565BA, Agilent technologies, Santa Clara, CA, US) and Feature Extraction software 10.7 (Agilent technologies, Santa Clara, CA, US) with default settings. Raw data were normalized by Quantile algorithm, Gene Spring Software 11.0 (Agilent technologies, Santa Clara, CA, US). Differentially expressed miRNAs were identified by using unpaired Student's t test with P values cutoff by 0.05 and fold change more than 2.0 or less than 0.5.
Target genes prediction, Gene ontology and pathway analysis
MicroRNA target gene prediction in gene ontology analysis was performed by miRTarBase Release 4.5 which is a public platform providing known experimentally validated miRNA targets37. GO analysis was applied to analyze the main function of the targets of differentially expressed miRNAs according to the Gene Ontology which is the key functional classification of NCBI38,39. GO analysis of target genes were performed by employing DAVID gene annotation tool40,41. Statistical analysis of GO terms was done by Fisher's exact test and x2 test and the false discovery rate (FDR) was calculated to correct the P-value,the smaller the FDR, the small the error in judging the p-value. The significant GO terms were defined as P value < 0.05 and FDR < 0.05. Likewise, pathway analysis was used to find out the significant pathway of target genes of differentially expressed miRNAs according to KEGG and Biocarta42. Still, we turned to the Fisher's exact test and x2 test to select the significant pathway and the threshold of significance was defined by P-value and FDR. The significant pathway was identified by P value < 0.05 and FDR < 0.05. We visualized the associations between target genes and miRNAs\pathways by using functional classification 2-D view analysis module of DAVID annotation tool. MiRNA-target interaction was analyzed by miRTar web server of human (http://mirtar.mbc.nctu.edu.tw/human/). We picked the miRNA-target interactions in biological pathways of interest and used Cytoscape for graphical representations.
Quantitative reverse transcription PCR
The qRT-PCR was performed on the CFX96 Touch™ (BIORAD, USA). The first strand of cDNA was synthesized with adjusted concentration of RNA and corresponding genes were amplified by employing EVA Green Supermix (BIORAD, USA). All the primers used for qRT-PCR were obtained from GeneCopoeia (USA).
Flow cytometry
The cells were collected by centrifugation at speed of 2000 rpm. The cell pellets were suspended with blocking buffer. After washing with blocking buffer, the cells were stained with corresponding mAbs for 30 min at room temperature. The monoclonal antibodies (MAb) used were either conjugated with fluorescein isothiocyanate (FITC), phycoerythrin (PE) or phycoerythrin-cyanin 5 (PerCP). Cell surface markers of FasL, NKG2D and TNFSF10 were labeled with PE conjugated mAb. CD40LG, CD94, KIR2DS5 and KIR2DL4 were stained by FITC conjugated mAb. 2B4 and NKG2C were stained with PerCP labeled mAbs against corresponding markers. After staining, the cells were washed twice before FACS analysis.
Western Blot
PBMCs, CIKIL-2 and CIKIL-15 were treated with cell lysis buffer and the concentration of total proteins extracted were measured by Lowry based method. The samples were analyzed by 12% SDS-PAGE gel loaded with equal amounts of protein. The proteins were electransferred to PVDF membrane at 40 V for 100 min. Next, the membrane was incubating with 5% skimmed milk in PBST for blocking overnight. The primary antibodies against TNF-α, PRF1, Bcl-2, GZMB, c-Myc, N-RAS and β-actin were added and incubated at room temperature for 4 hours. The HRP conjugated secondary antibodies were added after three time PBST washing. After incubating, the membranes were washed thoroughly with PBST for 4 times and then the bands were visualized by enhanced chemiluminescence kit (Millipore, USA).
References
Sawyers, C. L. et al. AACR Cancer Progress Report 2013. Clin Cancer Res. 19, S4–98 (2013).
Li, X. D. et al. Review of Chinese clinical trials on CIK cell treatment for malignancies. Clin Transl Oncol. 14, 102–108 (2012).
Stroncek, D. et al. Developments in clinical cell therapy. Cytotherapy. 12, 425–428 (2010).
Schmidt-Wolf, I. G., Negrin, R. S., Kiem, H. P., Blume, K. G. & Weissman, I. L. Use of a SCID mouse/human lymphoma model to evaluate cytokine-induced killer cells with potent antitumor cell activity. J Exp Med. 174, 139–149 (1991).
Jiang, J., Wu, C. & Lu, B. Cytokine-induced killer cells promote antitumor immunity. J Transl Med. 11, 83 (2013).
Lopez, R. D., Waller, E. K., Lu, P. H. & Negrin, R. S. CD58/LFA-3 and IL-12 provided by activated monocytes are critical in the in vitro expansion of CD56+ T cells. Cancer Immunol Immunother. 49, 629–640 (2001).
Jakel, C. E. & Schmidt-Wolf, I. G. An update on new adoptive immunotherapy strategies for solid tumors with cytokine-induced killer cells. Expert Opin Biol Ther. 14, 905–916 (2014).
Lin, G. et al. Interleukin-6 inhibits regulatory T cells and improves the proliferation and cytotoxic activity of cytokine-induced killer cells. J Immunother. 35, 337–343 (2012).
Wei, C. et al. The CIK cells stimulated with combination of IL-2 and IL-15 provide an improved cytotoxic capacity against human lung adenocarcinoma. Tumour Biol. 35, 1997–2007 (2014).
Wang, W. et al. Global transcriptome-wide analysis of CIK cells identify distinct roles of IL-2 and IL-15 in acquisition of cytotoxic capacity against tumor. BMC Med Genomics. 7, 49 (2014).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell. 136, 215–233 (2009).
Lewis, B. P., Burge, C. B. & Bartel, D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120, 15–20 (2005).
Sassen, S., Miska, E. A. & Caldas, C. MicroRNA: implications for cancer. Virchows Arch. 452, 1–10 (2008).
Jasinski-Bergner, S., Mandelboim, O. & Seliger, B. The role of microRNAs in the control of innate immune response in cancer. J Natl Cancer Inst. 106 (2014).
Xiao, C. et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 9, 405–414 (2008).
Fabbri, M. et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA. 305, 59–67 (2011).
Carson, W. E. et al. Interleukin (IL) 15 is a novel cytokine that activates human natural killer cells via components of the IL-2 receptor. J Exp Med. 180, 1395–1403 (1994).
Fehniger, T. A. & Caligiuri, M. A. Interleukin 15: biology and relevance to human disease. Blood. 97, 14–32 (2001).
Waldmann, T. A., Dubois, S. & Tagaya, Y. Contrasting roles of IL-2 and IL-15 in the life and death of lymphocytes: implications for immunotherapy. Immunity. 14, 105–110 (2001).
Leong, J. W., Sullivan, R. P. & Fehniger, T. A. microRNA management of NK-cell developmental and functional programs. Eur J Immunol. 44, 2862–2868 (2014).
Fehniger, T. A. et al. Next-generation sequencing identifies the natural killer cell microRNA transcriptome. Genome Res. 20, 1590–1604 (2010).
Nakagawa, M., Tsuzuki, S., Honma, K., Taguchi, O. & Seto, M. Synergistic effect of Bcl2, Myc and Ccnd1 transforms mouse primary B cells into malignant cells. Haematologica. 96, 1318–1326 (2011).
Martinez-Lorenzo, M. J. et al. Involvement of APO2 ligand/TRAIL in activation-induced death of Jurkat and human peripheral blood T cells. Eur J Immunol. 28, 2714–2725 (1998).
Thiery, J. & Lieberman, J. Perforin: a key pore-forming protein for immune control of viruses and cancer. Sub-cellular biochemistry. 80, 197–220 (2014).
Rousalova, I. & Krepela, E. Granzyme B-induced apoptosis in cancer cells and its regulation (review). Int J Oncol. 37, 1361–1378 (2010).
Elgueta, R. et al. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 229, 152–172 (2009).
Cretney, E., Takeda, K. & Smyth, M. J. Cancer: novel therapeutic strategies that exploit the TNF-related apoptosis-inducing ligand (TRAIL)/TRAIL receptor pathway. Int J Biochem Cell Biol. 39, 280–286 (2007).
Loskog, A. & Totterman, T. H. CD40L - a multipotent molecule for tumor therapy. Endocrine, metabolic & immune disorders drug targets. 7, 23–28 (2007).
Lauber, S. et al. Novel function of Oncostatin M as a potent tumour-promoting agent in lung. Int J Cancer. 136, 831–843 (2015).
West, N. R., Murray, J. I. & Watson, P. H. Oncostatin-M promotes phenotypic changes associated with mesenchymal and stem cell-like differentiation in breast cancer. Oncogene. 33, 1485–1494 (2014).
Fossey, S. L., Bear, M. D., Kisseberth, W. C., Pennell, M. & London, C. A. Oncostatin M promotes STAT3 activation, VEGF production and invasion in osteosarcoma cell lines. BMC Cancer. 11, 125 (2011).
Viant, C. et al. SHP-1-mediated inhibitory signals promote responsiveness and anti-tumour functions of natural killer cells. Nature communications. 5, 5108 (2014).
Moretta, L. et al. Human natural killer cells: origin, receptors, function and clinical applications. International archives of allergy and immunology. 164, 253–264 (2014).
Le Drean, E. et al. Inhibition of antigen-induced T cell response and antibody-induced NK cell cytotoxicity by NKG2A: association of NKG2A with SHP-1 and SHP-2 protein-tyrosine phosphatases. Eur J Immunol. 28, 264–276 (1998).
Cheent, K. S. et al. Synergistic inhibition of natural killer cells by the nonsignaling molecule CD94. Proc Natl Acad Sci U S A. 110, 16981–16986 (2013).
Borrego, F., Ulbrecht, M., Weiss, E. H., Coligan, J. E. & Brooks, A. G. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med. 187, 813–818 (1998).
Hsu, S. D. et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 39, D163–169 (2011).
The Gene Ontology (GO) project in 2006. . Nucleic Acids Res. 34, D322–326 (2006).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25, 25–29 (2000).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 4, 44–57 (2009).
Dennis, G., Jr et al. DAVID: Database for Annotation, Visualization and Integrated Discovery. Genome Biol. 4, P3 (2003).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Acknowledgements
We thank Dr. Zhu Xiaoyun for his constructive suggestions to this study. This work is supported by grants from National Natural Science Foundation of China (No. 81160267, 81360245 and 81460436) and grants from the “Special and Joint Program” of Yunnan Province Science and Technology Department & Kunming Medical University.
Author information
Authors and Affiliations
Contributions
Z.L.H. and W.J.W. conceived and designed the study. W.J.W., M.Y.M. and Y.Y.Z. performed the experiments. W.J.W., R.H.L. and R.Y.D. analyzed the data. C.H.W., B.H.L. and W.W.T. contributed reagents/material. C.Y.W., Y.H.X. and L.H.J. interpreted the data and wrote the paper. F.Y., Y.M.Z. and X.F.J. revised the manuscript. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, W., Li, R., Meng, M. et al. MicroRNA profiling of CD3+CD56+ cytokine-induced killer cells. Sci Rep 5, 9571 (2015). https://doi.org/10.1038/srep09571
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep09571
This article is cited by
-
Dysregulation of micro-RNA 143-3p as a Biomarker of Carotid Atherosclerosis and the Associated Immune Reactions During Disease Progression
Journal of Cardiovascular Translational Research (2024)
-
microRNA-143-3p contributes to inflammatory reactions by targeting FOSL2 in PBMCs from patients with autoimmune diabetes mellitus
Acta Diabetologica (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.