Abstract
CCT domain-containing genes generally control flowering in plants. Currently, only six of the 41 CCT family genes have been confirmed to control flowering in rice. To efficiently identify more heading date-related genes from the CCT family, we compared the positions of heading date QTLs and CCT genes and found that 25 CCT family genes were located in the QTL regions. Association mapping showed that a total of 19 CCT family genes were associated with the heading date. Five of the seven associated genes within QTL regions and two of four associated genes outside of the QTL regions were confirmed to regulate heading date by transformation. None of the seven non-associated genes outside of the QTL regions regulates heading date. Obviously, combination of candidate gene-based association mapping with linkage analysis could improve the identification of functional genes. Three novel CCT family genes, including one non-associated (OsCCT01) and two associated genes (OsCCT11 and OsCCT19) regulated the heading date. The overexpression of OsCCT01 delayed flowering through suppressing the expression of Ehd1, Hd3a and RFT1 under both long day and short day conditions. Potential functions in regulating heading date of some untested CCT family genes were discussed.
Similar content being viewed by others
Introduction
The transition from vegetative to reproductive growth directly determines flowering time in plants. Among monocots, the long-day plants barley and wheat require complete vernalization (a long period with low temperature and long day length) before flowering1. In contrast, flowering in rice, a short-day plant and maize, either a day-neutral or a short-day plant, depends on a short day length2,3,4.
The molecular mechanisms underlying flowering have been extensively studied in Arabidopsis. It is well known that a circadian clock–controlled flowering pathway comprising the genes GIGANTEA (GI), CONSTANS (CO) and FLOWERING LOCUS T (FT) promotes flowering under long day conditions (LD)5,6,7. That is, GI regulates circadian rhythms and CO expression and CO upregulates FT expression and results in early flowering under LD8,9. This GI-CO-FT regulatory pathway is conserved in rice. Under LD, OsGI regulates the expression of Heading date 1 (Hd1), the ortholog of Arabidopsis CO4. Hd1 negatively regulates Heading date 3a (Hd3a), the ortholog of Arabidopsis FT and delays the heading date10. Hd1 has a dual function in regulating flowering, as it suppresses flowering under LD but promotes flowering under short-day conditions (SD) by influencing the expression of Hd3a4,11. A specific flowering pathway mediated by Ehd1-Hd3a was also identified in rice2. In this rice-specific pathway, Ehd1 upregulates the expression of Hd3a and promotes flowering under LD. OsGI is required for induction of Ehd1 and Hd3a. OsGI-Ehd1 is more dominant for flowering-time control than OsGI-Hd1 in rice12,13.
In the photoperiod flowering pathway, CO belongs to gene family encoding CCT (CO, CO-LIKE and TOC1) domain proteins. The CCT domain was originally described as a 43 amino acid-long region of homology found in the Arabidopsis thaliana proteins CONSTANS (CO), CO-LIKE and TIMING OF CAB1 (TOC1), which have been well characterized as regulators of flowering in Arabidopsis14,15,16. CCT family members could be classified into three sub-classes, COL, CMF (CCT MOTIF FAMILY) and PRR (PSEUDORESPONSE REGULATOR) according to their motifs17. COLs contain one CCT domain and B-boxes. CMF genes contain only one CCT domain. PRRs share a conserved domain, the receiver-like domain (RLD), along with a CCT domain. The RLD is similar to the receiver domain of the response regulators in the histidine to aspartic acid (His–Asp) phosphorelay, a versatile signal transduction system18. A combined inter-/intra-species comparative and phylogenetic analysis of the CMF, COL and PRR gene families in Poaceae indicates that they evolved prior to the monocot/dicot divergence approximately 200 mya17.
Several studies have indicated that CCT domain-containing genes are key flowering regulators in rice. Ghd7, a CMF class gene, functions upstream of Ehd1 and Hd3a in the photoperiod flowering pathway19. OsCO3, a COL gene, controls flowering by negatively regulating the expression levels of FT-like genes under SD conditions, resulting in earlier flowering20. OsCOL4, a constitutive flowering repressor, acts upstream of Ehd1 and downstream of OsphyB21. DTH2, a COL class gene, promotes heading by inducing Hd3a and RICE FLOWERING LOCUS 1 (RFT1) under LD22. Ghd7.1 encodes a PRR37-like protein that represses flowering by suppressing the expression of the downstream genes Ehd1 and Hd3a under LD, greatly enhancing grain productivity23. All these studies indicated that the members in each CCT subfamily have potential roles in regulating the heading date. In other cereals, several flowering regulators were identified, including HvCO1 and HvCO9, the homologues of Hd1 and Ghd7 in barley24,25; Ppd-H1, Ppd-D1 and SbPRR37, the homologues of Ghd7.1 in barley, wheat and sorghum, respectively26,27,28; and ZmCCT, the homologue of Ghd7 in maize3 and VRN2 in wheat29. These genes are all CCT family genes that regulate flowering, which indicates that some genes controlling flowering are conserved at the level of the DNA sequence and share a biological function within cereals, although these cereals have very different growth behaviors and growth regions.
In the last two decades, hundreds of QTLs have been reported for the heading date (http://www.gramene.org/). The major QTLs, such as Hd1, Ghd7 and Ghd7.1, have been identified by a map-based cloning strategy4,19,23. All these QTLs contain a CCT domain and contribute to rice grain yield through extending the heading date under LD. In rice, it was reported that there are 36 candidate genes in the CCT family, among which 17 genes belong to OsCOL, 14 genes belong to OsCMF and five genes belong to OsPRR17. Although quite a few CCT genes have been characterized for heading date, the functions on flowering of the major members in this family remain unclear.
Association mapping is an efficient alternative to establish the relationships between genes and traits. The major flowering QTL, Vegetative to generative transition 1, was associated with flowering and then confirmed by genetic engineering30. Based on a diverse panel of Chinese germplasm of rice, several marker loci on chromosome 7 were repeatedly associated with agronomical traits including heading date31. In recent years, next-generation sequencing has greatly promoted genome-wide association mapping in plants. Genome-wide association mapping for 14 agronomic traits were conducted based on a population of 373 indica lines. Seven genes were associated with heading date32. Candidate gene-based association mapping were widely used in maize. For example, one polymorphism in the fifth exon of ZmGS3 is significantly associated with kernel length in two environments33. ZmCCT, encoding a CCT domain-containing protein, is associated with the photoperiod response3. Gene family is an important resource of candidate gene-based association mapping. The association between ZmDREB genes and drought tolerance was evaluated using a diverse population of maize consisting of 368 varieties from tropical and temperate regions34. A significant association between the DNA polymorphisms in the promoter region of ZmDREB2.7 and drought tolerance at seedling stage was identified and further confirmed by transgenic lines in Arabidopsis. But flowering candidate gene-based association mapping was not reported.
In this study, to efficiently identify heading date genes from the CCT family, we first searched for all CCT family members and compared their genomic positions with QTLs for heading date. Then we identified the CCT members with potential functions in regulating heading date via a candidate gene-based association analysis. Finally, we tested the potential functions of 18 candidate CCT members by genetic transformation. Our purpose is to claim how many novel CCT genes regulate rice heading and whether associated CCT genes located in the heading date QTL regions have more chance involved in heading.
Results
Comparison of genome positions between CCT family genes and heading date QTLs
We previously identified a total of 41 CCT family genes in the rice genome35. They were mapped to 12 chromosomes according to their physical positions in the genome and included 35 genes but not LOC_Os08g02620, which was reported by Cockram et al.17 (Fig. 1, Table S2). These genes are unevenly distributed on all 12 chromosomes. Only one CCT family gene is situated on chromosomes 1 and 4, while eight genes are located on chromosome 2. The remaining chromosomes each harbor two to seven CCT family genes. OsCCT03, OsCCT04, OsCCT24 and OsCCT25, OsCCT36 and OsCCT37, OsCCT39 and OsCCT40 were closely linked to each other, within 300 kb. Accordingly, 133 rice heading date QTLs were collected in the Gramene web site (http://www.gramene.org/, release 37). Finally, a total of 59 heading date QTLs were merged into 12 chromosomes by integrating closely linked QTLs (Fig 1, Table S5). We located 41 CCT family genes and 59 integrated QTLs on 12 chromosomes according to their physical positions by a program compiled by R language (http://www.r-project.org/). Of them, 13 QTLs were isolated, including OsHY2, DTH2, Ehd4, Hd6, Hd16, Hd17, RFT1, Hd3a, Hd1, Ghd7, Ghd7.1, Ghd8 (also known as DTH8 and Hd5) and Ehd12,4,10,19,22,23,36,37,38,39,40,41,42. Chromosome 1 carried the largest number of eight QTLs. On the contrary, chromosome 12 carried the smallest number of three QTLs. A comparison of the genomic positions of the CCT genes with the heading date QTLs showed that 25 CCT family genes were located in the regions of the heading date QTLs. The remaining 16 CCT genes were located outside of heading date QTL regions (Fig 1). Conversely, 39 heading date QTL regions do not contain CCT genes. Among the 13 cloned heading date QTLs, Qhd14 (DTH2), Qhd31 (Hd1), Qhd36 (Ghd7) and Qhd39 (Ghd7.1) are CCT genes, which corresponded to OsCCT08, 21, 26 and 28 in this study. Interestingly, chromosome 1 carried the largest number of heading date QTLs but the smallest number of CCT genes. In contrast, chromosome 2 carried the largest number of heading date QTLs and CCT genes.
Phylogenetic tree of CCT family genes
To understand the evolution of the CCT family genes, protein sequences deduced from Nipponbare were used to construct a neighbor-joining phylogenetic tree based on the conserved CCT domain, the B-box (COL) and the REC domain (PRR). The CCT family was clearly resolved to three subfamilies corresponding to CMF, COL and PRR (Figure 2). These genes included 19 CMF genes, 17 COL genes and five PRR genes (Table S2). OsCCT26 (Ghd7), a CMF subfamily gene, delayed flowering by up to 21 days under LD19. OsCCT28 (Ghd7.1), a PRR subfamily gene, delayed flowering by 14 days under LD23. OsCCT08 (DTH2), a COL subfamily gene, promoted flowering by eight days under LD22. OsCCT35, the gene evolutionarily closest to OsCCT26 (Ghd7), is located in the region of Qhd51 on chromosome 10. OsCCT11, the gene evolutionarily closest to OsCCT28 (Ghd7.1), is located in the region of Qhd16 on chromosome 3. OsCCT22, the closest gene to OsCCT08 (DTH2), is located on Chromosome 6.
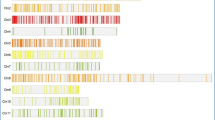
Phylogenetic tree of rice CCT family genes and their expression profiling in leaves and stems.
The phylogenetic tree was constructed based on the amino acid sequences of the proteins. The expression data was extracted from a database (http://ricexpro.dna.affrc.go.jp). The expression data on the 6 genes in red were missing. T1: Leaf blade_27DAT_12:00; T2: Leaf blade_76DAT_12:00; T3: Leaf blade_125DAT_12:00; T4: Stem_83DAT_12:00; T5: Stem_90DAT_12:00.
Expression profiling of CCT family genes
Rice heading is largely regulated by photoperiod. Light is mainly perceived by leaves and stems before heading. Therefore, we extracted expression data on 35 CCT family genes generated from leaves and stems in the japonica rice Nipponbare (http://ricexpro.dna.affrc.go.jp). In general, the expression levels varied greatly among genes (Figure 2). In the CMF subfamily, four genes (OsCCT03, 04, 13 and 18) had high expression levels in leaves and stems, two genes (OsCCT15 and 35) had considerable expression levels in leaves but were low in stems, seven genes had very low expression levels in all tested tissues and five genes failed to generate expression data. In the COL subfamily, seven genes, including COL4, were highly expressed in all tested tissues and the other three genes, DTH2, CO3 and OsCCT05, were highly expressed only in leaves; four genes, Hd1, OsCCT09, 30 and 32, had very low expression levels. The PRR subfamily genes, except OsCCT07, had higher expression levels in leaves than in stems.
Nucleotide diversity of CCT genes
A total of 2,066 SNPs were identified in 41 CCT family genes from 529 accessions. Of these SNPs, 284 are located in exons and 662 are located in introns. Other SNPs are located in promoters (943), 5′ UTRs (47) and 3′ UTRs (130). The level of natural variation differed among genes (Table S6). The pairwise nucleotide diversity ranged from one to eight SNPs per kilobase for most CCT genes. The average nucleotide diversity of CCT family genes (π = 2.6 × 10−3) is equivalent to that of the whole Oryza sativa genome (π = 2.4 × 10−3)43. OsCCT18 had the lowest diversity with only one SNP per 10 kilobases in O. sativa and OsCCT29 had the highest diversity with approximately eight SNPs per kilobase in both O. sativa and wild rice. Tajima's D of 16 CCT genes, OsCCT05/08/09/10/13/16/19/21/23/25/28/30/31/35/38/41 (P < 0.01), seven CCT genes, OsCCT01/15/17/20/22/24/26 (P < 0.001), reached a significant positive level. The ratio πc/πw for CCT12, 16 and 18 was less than 0.5, indicating that these genes encountered selection during domestication and genetic improvement (Table S6). In particular, CCT18 has a very low ratio πs/πw of 0.03, indicating a 34-fold reduction in genetic variation in O. sativa.
Association mapping for heading date
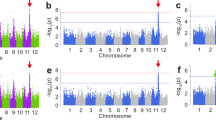
The 529 accessions with a wide range of heading dates were used to examine associations between CCT family genes and heading date. 529 accessions were classified into five subgroups indica 1 (Ind1), indica 2 (Ind2), an admixture of indica (Ind), tropical japonica (TrJ), temperate japonica (TeJ) and an admixture of japonica (Jap) (Figure S1). No associations were detected between the 41 CCT family genes and heading date in both the whole indica group and whole japonica group. Then association mapping was conducted within subgroups in case of noise caused by population structure. No associations were detected either in TrJ or Jap subgroups in both years. A total of 19 and 16 CCT family genes were associated with heading date at P < 0.001 in 2012 and 2013, respectively. Except OsCCT10, OsCCT11 and OsCCT35 detected in 2012, associations of the other 16 genes with heading date were detected in both years (Table 1). Among them, five genes are the previously cloned genes and 10 genes are located in QTL regions. Seven, eight and three CCT family genes were significantly associated with heading date within the subgroups Ind1, Ind2 and Ind in 2012, respectively. In the TeJ subgroup, six and six genes were associated with heading date in 2012 and 2013, respectively. Of them, the associations of OsCCT07, 12 and 33 with heading date were only detected in the TeJ subpopulation in 2012. But in 2013, the association of OsCCT31 with heading date was detected only in TeJ subgroup. The associations of six OsCCT genes with heading date were commonly detected in the TeJ subgroup and at least one indcia subgroup in 2012 or 2013. The associations of OsCCT28 were detected in three subgroups in 2012 and four subgroups in 2013. Among all the associated genes, OsCCT06 showed the highest significant association at P = 9.4 × 10−7 in 2012. In 2013, OsCCT28 showed the highest significant association at P = 5.2 × 10−12. The heading date-associated genes OsCCT08, OsCCT26 and OsCCT28 corresponded to the heading date QTLs DTH2, Ghd7 and Ghd7.1, respectively, which were identified in bi-parental populations. OsCCT06 and OsCCT31 are CoL4 and Co3, respectively, which were reported as repressors of flowering due to the phenotypes of col4 mutants and Co3-overexpressing plants20,21. The non-synonymous SNP C/A in the first exon of CoL4 is associated with heading date. More specifically, the average heading date (111 d) of the C genotype was much later than that (92 d) of the A genotype (P = 1.3 × 10−7). However, the major QTL Hd1 (OsCCT21) was not associated with heading date in any group or subgroup.
Functional validation of CCT family genes by transformation
In total, 41 CCT family genes were classified into four groups according to their associations with heading date and genomic positions. Genes in group 1 were associated with heading date and fell into heading date QTL regions. This group includes 12 genes, including OsCCT08 and OsCCT09. All cloned heading date QTLs/genes belonging to the CCT family except OsCCT21 (Hd1) are classified into group 1. Genes in group 2 fell into heading date QTL regions but were not associated with heading date. There are 14 genes in group 2 such as OsCCT01. Genes in group 3 were associated with heading date but not contained in heading date QTL regions. They comprise six genes, including OsCCT12 and OsCCT13. Genes in group 4 were not associated with heading date or contained in heading date QTL regions. There are nine genes in group 4, including OsCCT02 and OsCCT22. To clarify whether genes in different groups have different likelihoods of regulating heading date, we chose a total of 18 genes, including four in group 1, five in group 2, two in group 3 and seven in group 4 to test their functions on heading by transformation (Table 2). We generated transgenic plants overexpressing or silencing each of 18 CCT genes (Figure S2 and Figure S3), among which nine genes were amplified from japonica Nipponbare and nine genes were amplified from the late-flowering indica cultivar Minghui 63 (Table S4) using gene-specific primers (Table S3). The transgenic plants carrying all nine tested genes in groups 3 and 4 had no altered heading date (Table 2). Among the four tested genes in group 1, OsCCT11 and OsCCT19 had effects on heading date. Transgenic plants overexpressing OsCCT19 (Figure S3) exhibited delayed heading by up to five days and the OsCCT11 RNAi plants (Figure S3) delayed heading by up to 12 days under LD (Table 3). The other two tested genes, OsCCT09 and OsCCT10, had no effects on heading. Among the five tested genes in group 2, overexpression of OsCCT01 (Figure S3) greatly delayed heading by more than 25 days but resulted in a decreased plant height and short panicles (Fig 3). The other four genes had no effects on heading when overexpressed.
Expression pattern of OsCCT01 and the subcellular localization of OsCCT01
We investigated the diurnal expression pattern of OsCCT01 in young leaves of Zhonghua 11 using qRT-PCR (Fig S4). The OsCCT01 transcript was more abundant during the day than in darkness. The expression of OsCCT01 was low under SD, while its expression peaked at similar levels in the morning under both SD and LD. To assay the sub-cellular localization of OsCCT01, the coding sequence of OsCCT01 was fused to that of yellow fluorescent protein (YFP) and Ghd7, a nuclear protein, was fused to cyan fluorescent protein (CFP). The constitutive 35S cauliflower mosaic virus promoter was used to drive expression of the two fused fluorescent proteins. The constructs were co-transfected into rice protoplasts derived from etiolated seedlings. The merged picture showed that OsCCT01 co-localized with GHD7, indicating that OsCCT01 is a nuclear protein (Figure S5).
Photoperiod sensitivity of OsCCT01
To investigate the function of OsCCT01 in the photoperiodic control of flowering, the T2 generation of transgenic plants was used to examine flowering time under LD and SD (10 h light and 14 h darkness) in a natural field. OsCCT01-overexpressing plants exhibited 14 day-later flowering under SD and 25 day-later flowering under LD relative to the negative control plants (Table 4). These results suggest that OsCCT01 regulates flowering time in rice under both SD and LD. We compared the expression of OsCCT01 and other key flowering regulators in young leaves between wild-type Zhonghua 11 and OsCCT01-overexpressing plants using qRT-PCR. The expression levels of Ehd1, Hd3a, and RFT1 were downregulated in the transgenic lines compared to the wild-type plants under both SD and LD. In contrast, the transcript levels of Hd1 were not obviously affected (Figure 4). The expression of other flowering regulators such as OsGI, RID1/OsID1/Ehd2, Ehd3, Ehd4, OsMADS50, OsMADS51, OsMADS56, Ghd7, Ghd8/DTH8, Ghd7.1 and DTH2, was also not affected by OsCCT01 (Figure S6 and S7). Thus, we deduced that OsCCT01 acted as a negative regulator upstream of Ehd1 in the photoperiod flowering pathway under SD and LD.
Discussion
The extent of linkage disequilibrium determines the resolution of association mapping and the number of markers required for genome-wide association analysis44,45. Candidate gene-based association analysis has the potential to identify putative functional genes in several species including humans, animals and maize, in which quick linkage disequilibrium decay has been reported46,47,48. Candidate genes frequently come from some gene families, which have been characterized as associating with target traits, from genes involving targeted biosynthesis pathways, such as the tocopherol biosynthesis genes49 and from the potential genes in small genome regions delimited by map-based cloning40.
Many crucial flowering-related rice genes contain the CCT domain, such as Hd1, Ghd7, Ghd7.1 and DTH24,19,22,23. Naturally, the other CCT family genes were regarded as candidate genes for association analysis. A total of 19 CCT genes, including five previously cloned flowering-related genes, were associated with heading date. Of these, six were analyzed for functions by transformation. The regulation of heading date by OsCCT11 and OsCCT19 was further confirmed by RNAi and overexpression, respectively. Both genes are located in QTL regions. The other four genes (OsCCT09, OsCCT10, OsCCT33 and OsCCT41) have no effects on heading date. We tested 12 CCT genes not associated with heading date by transformation. Overexpression of one gene (OsCCT01) significantly delayed heading. Moreover, an analysis of the expression levels of several key flowering-related genes showed that OsCCT01 suppressed Ehd1 and further suppressed Hd3a under either SD or LD, resulting in delayed heading. Therefore, OsCCT01 is a flowering suppressor. The effects of OsCCT01 on plant height and grain yield are distinct from the cloned CCT family genes such as Ghd7 and Ghd7.1. Both Ghd7 and Ghd7.1 increase plant height and grain yield19,23, whereas OsCCT01 decreases plant height and grain yield mainly by decreasing the length of stem internodes, the number of spikelets per panicle and seed setting rate (Table S8).
There are nine CCT family genes that regulate heading date if including the previously cloned six genes. Among the nine functional genes, five genes were located within QTL regions and two genes were located outside of QTL regions and were associated with heading date; two genes located in the QTL regions were not associated with heading date. In summary, five of seven associated genes in the QTL regions have effects on heading date, two of six non-associated genes in the QTL regions regulate heading date, two of four associated genes outside of QTL regions regulate heading date and none of the seven non-associated genes situated outside of the QTL regions regulates heading date. Therefore, the combination of association mapping with linkage analysis improved the identification of functional genes.
Seven of the nine functional genes, all except Hd1 and OsCCT01, were detected by association mapping. Thus, candidate gene-based association analysis has considerable power in QTL mapping. However, four of the 11 associated genes have no effect on heading date as confirmed by either gene overexpression or gene silencing and two (OsCCT21/Hd1 and OsCCT01) functional genes were not associated with heading date. That result indicates that type I error and type II error occurred. Type I error is frequently caused by low threshold values. Moreover, low frequency alleles were excluded when conducting association mapping. Thus, it is hard to avoid type II error. In fact, only approximately one half of the heading date-related genes were found to be associated with heading date even using a large number of landraces43. In this study, the most possible reason for type I error is a large degree of linkage disequilibrium in the target regions43 and a close linkage between tested genes and a single heading date gene in the linkage disequilibrium regions. For example, OsCCT09 and OsCCT10 are closely linked to DTH2 and EHD4. In addition, it is also possible that functional redundancy exists among CCT genes50. Silencing some genes might not cause any phenotypic change. As for OsCCT01 and OsCCT19, changes in heading date were observed in the overexpression plants, but no change was observed in the RNAi plants. Thus, the functions of these CCT family genes (OsCCT09, 10, 29 and 38) tested by RNAi are uncertain. Knockout mutations created by the high efficiency gene-editing method Clustered Regularly Interspaced Short Palindromic Repeat (CRISPRS)/CRISPR-associated (Cas) systems51 would be much better than knockdown mutations for testing the functions of family genes.
A negative Tajima's D signifies an excess of low frequency polymorphisms relative to expectations, indicating a selective sweep or purifying selection. A positive Tajima's D signifies balancing selection. The Tajima' D test (P < 0.01) indicated that no significant Tajima's D was detected in wild rice. However, in cultivar, Tajima's D values for OsCCT18 were negative and significant. Clearly, OsCCT18 was selected during its natural evolutionary process. However, we did not observe any function of OsCCT18 in the overexpression plants. Therefore, we examined the DNA polymorphisms in eight noncoding regions (ranging from 313 bp to 2,055 bp) flanking OsCCT18 (Table S4) because noncoding regions eliminate the interference of other genes' variation. We observed a selective sweep with a notable reduction in relative diversity in a small segment (< 50 kb) (Table S7). These results indicated that a strong selection pressure was imposed on the region surrounding OsCCT18 during rice domestication. Therefore, OsCCT18 has no obvious phenotypic function but infrequent selection. Tajima's D values for OsCCT01, 17, 20 and 24 were positive and significant. Huang et al.43 reported that more than 60 genome regions experienced infrequent selection in cultivars compared to wild rice. Of them, the selected regions (19.9–21.1 Mb in indica/22.8–22.9 Mb in japonica and 29.7–29.9 Mb) contained OsCCT17 and 18. It is likely that directional selection on favorable alleles in the regions caused genetic hitchhiking effects on linked OsCCT17 and 18.
In this study, in addition to the previously cloned six CCT family genes, three CCT family genes were identified as regulating heading date. We believe that more genes regulate heading date because the functions of the other eight genes associated with heading date remain to be confirmed by transformation. In addition, the functions of two CCT family genes (OsCCT33 and 41), which were associated with heading date, were likely misevaluated by RNAi because of gene redundancy. A combination of overexpression and gene knockout methods to test gene function would achieve more reliable results. Currently, it is not clear whether OsCCT20 and OsCCT24 regulate heading date. The Tajima test indicated that both genes underwent balancing selection, but the regions surrounding both genes were not reported to suffer selection. Therefore, genetic hitchhiking effects could not be used to explain their infrequent selection; most likely, they are heading date-related genes. OsCCT11 is the member closest to the major heading date-related gene OsCCT28 (Ghd7.1) and OsCCT21 (Hd1) is close to OsCCT31 (CO3). Both genes regulate heading date. OsCCT35 and OsCCT16 are the members closest to the key heading date genes OsCCT26 (Ghd7) and OsCCT06 (COL4), respectively. OsCCT35 and OsCCT16 have potential functions in flowering. These CCT family genes should be tested for their functions.
Methods
Measurement of heading date
The Chinese core collection consisting of 202 cultivars and the world core collection of 327 cultivars and an additional 107 common wild rice accessions (Oryza rufipogon) were sowed at the farm of Huazhong Agricultural University Wuhan, China on May 19 2012 and on April 17 2013. The basic information for 529 cultivars is available on the RiceVarMap52 (http://ricevarmap.ncpgr.cn/) and the details of the 107 wild rice accessions are included in Table S1. Seven 25-day-old seedlings from each accession were transplanted in a single row with a distance of 16.5 cm between plants and 26.4 cm. Field management was performed according to normal agricultural practices. The heading date was defined as the number of days from sowing to the appearance of the first panicle. The 5 plants in the middle were used to score the heading date. The average heading date of the 5 plants was used for data analysis.
Integration of heading date QTLs
The key words “heading date in rice” and “flowering in rice” were used as queries to search against the annotation database of the rice genome (http://www.gramene.org/, release 37). Then, the physical positions of all the collected QTLs were obtained through aligning the sequences of each QTL's flanking RFLP marker to the rice reference genome (Gramene Annotated Nipponbare Sequence 2009) by BLAST (BLAST-like alignment search tool)53 or performing an e-PCR54 using the pair of primers of corresponding SSR markers to the whole genome sequence55. Considering a 1-LOD confidence interval of more than 5 cM for a QTL in a primary mapping population, each couple of QTLs with a physical distance less than 1250 kb (approximately 5 cM) were merged into one.
Identification of CCT family genes in rice
The conserved CCT domain sequence EREAKLMRYKEKRKKRCYEKQIRYASRKAYAEMRPRVRGRFAKE of the known GHD7 protein was used to search (using the tBLASTn and BLASTp programs) against the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/)/NCBI (http://www.ncbi.nlm.nih.gov)/DRTF (http://drtf.cbi.pku.edu.cn/index.php)35. A total of 41 CCT genes were identified and named sequentially according to their genome positions (Table S2). The unrooted phylogenetic tree of CCT family members was constructed with the MEGA5.2.2 program by the neighbor-joining method and bootstrap analysis (1000 replicates)56. The expression pattern heat map of the CCT family members was drawn by the R language and expression profiling of 35 members came from RiceXPro (http://ricexpro.dna.affrc.go.jp).
Nucleotide diversity analysis
The genome sequences of the 529 accessions were downloaded from RiceVarMap (http://ricevarmap.ncpgr.cn/). The sequence of each CCT family gene was extracted from the dataset. Each sequence included the gene body, approximately 2 kb of the promoter region and approximately 500 bp of the 3′ flanking region. The average number of nucleotide differences per site between two random sequences (π), the Watterson estimator (θw), nucleotide diversity and Tajima's D statistics were calculated using the DnaSP 5.0 program57. The value of πc/πw was calculated with the average proportion of pairwise difference per base pair in cultivated rice (πc) divided by that in wild rice (πw).
Candidate gene-based association mapping
The population structure of 529 accessions was considered when conducting the association analysis. The parameter of the number of ancient clusters K was set from two to seven to obtain different inferences. The highest likelihood for a subpopulation was obtained with K = 652 (http://ricevarmap.ncpgr.cn). Associations between SNPs in CCT genes and heading date were analyzed with a general linear model (GLM) using the software TASSEL58. The Bonferroni-adjusted significance threshold is set as 0.05/n (n polymorphism)59. For most candidate genes, n is fluctuated from 40 to 60 with an average of approximate 50 within each subgroup. Hence P<0.001 was set the threshold for claiming association. Analyses were conducted with population structure estimates, using the Q-matrix obtained from the RiceVarMap (http://ricevarmap.ncpgr.cn).
Complementation test of heading date-related CCT members
Genome DNA fragments or coding DNA sequences of 18 CCT genes were amplified from Minghui 63 or Nipponbare DNA with gene-specific primers (Table S3) using LA Taq polymerase (Takara) with high fidelity. The transformation information for 18 CCT genes is listed in Table S4. The amplicons without mutations were identified by sequencing and inserted into the binary vectors pCAMBIAI1301s and ds1301. The resulting constructs were introduced into Zhonghua 11 or Hejing 19 by Agrobacterium tumefaciens-mediated transformation using immature embryos as subjects60. T1 and T2 families were sown in the field bed on 16 May and 30 plants of T1 families for each candidate gene were transplanted on 15 June 2011 and 2012 in the Experimental Station of Huazhong Agricultural University, Wuhan, China. The wild-type accessions Zhonghua 11 and Hejiang 19 were grown as the controls. Seven OsCCT01 overexpressed plants and seven negative plants were treated in the short day (10 h day length) and long day (14 h day length) conditions till heading respectively.
RNA extraction and expression analysis
The seeds of negative and positive OsCCT01 transgenic plants and wild-type were sown in pots in a greenhouse. After 32 d of growing under approximately neutral day-length conditions (12 h of light per day), half of the plants were moved to a short day condition (10 h/d) in a phytotron and the other half were moved to a long-day condition (14 h/d) in a different phytotron. The light intensity in the phytotrons was set at 10,000 lx, with the temperature set to 30°C in the light period and 26°C in the dark period. After an additional seven days of growth, young leaves were simultaneously harvested from three different plants as three biological repeats for each treatment and stored in liquid nitrogen. To test their circadian expression patterns, the samples were collected once every four hours for 24 hours. In addition, young fresh leaves from other CCT genes' transgenic plants were separately sampled for RNA extraction at seedling stage. 10 day seedling and 4 cm young panicle were sampled from negative and positive OsCCT01 transgenic plants for RNA extraction to detected rice yield genes and plant height involved genes, such as GAs.
Total RNA was extracted from the leaves using an RNA extraction kit (TRIzol reagent, Invitrogen) for quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR). Approximately 3 μg total RNA was reverse-transcribed using M-MLV reverse transcriptase (Invitrogen) in a volume of 150 μl to obtain cDNA. We investigated the transcription levels of 18 CCT genes in their corresponding transgenic plants and photoperiod genes including Hd1, Ehd1, Hd3a and RFT1 in OsCCT01 transgenic plants. Ubiquitin was set as the internal control (LOC_Os03g13170). All the primers used for qRT-PCR are presented in Table S3. qRT-PCR was run in a total volume of 15 μl containing 3.6 μl of the reverse-transcribed product described above, 0.25 μM gene-specific primers and 7.8 μl FastStart Universal SYBR Green Master (Rox) superMIX (Roche, Mannheim, Germany) on an Applied Biosystems ViiA 7 Real-Time PCR system according to the manufacturer's instructions. The measurements were obtained using the relative quantification method.
Expression analysis and sub-cellular localization
The coding sequence of OsCCT01 (MH63) was fused to PM999-YFP. The fusion protein with insertion in the correct direction was co-transfected into rice protoplasts with Ghd7:CFP as described by Zhou61, with minor modifications. The fluorescence image was obtained using a confocal microscope (Leica, Germany) after incubating the transformed cells in the dark at 28°C for 20 h.
References
Trevaskis, B., Hemming, M. N., Dennis, E. S. & Peacock, W. J. The molecular basis of vernalization-induced flowering in cereals. Trends in Plant Science 12, 352–357 (2007).
Doi, K. et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes & Development 18, 926–936 (2004).
Yang, Q. et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proceedings of the National Academy of Sciences 110, 16969–16974 (2013).
Yano, M. et al. Hd1, a Major Photoperiod Sensitivity Quantitative Trait Locus in Rice, Is Closely Related to the Arabidopsis Flowering Time Gene CONSTANS. The Plant Cell Online 12, 2473–2483 (2000).
Imaizumi, T. & Kay, S. A. Photoperiodic control of flowering: not only by coincidence. Trends in plant science 11, 550–558 (2006).
Kobayashi, Y. & Weigel, D. Move on up, it's time for change—mobile signals controlling photoperiod-dependent flowering. Genes & Development 21, 2371–2384 (2007).
Turck, F., Fornara, F. & Coupland, G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594 (2008).
Fowler, S. GIGANTEA : a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18, 4679–4688 (1999).
Park, D. H. et al. Control of Circadian Rhythms and Photoperiodic Flowering by the Arabidopsis GIGANTEA Gene. Science 285, 1579–1582 (1999).
Kojima, S. et al. Hd3a, a Rice Ortholog of the Arabidopsis FT Gene, Promotes Transition to Flowering Downstream of Hd1 under Short-Day Conditions. Plant and Cell Physiology 43, 1096–1105 (2002).
Hayama, R., Yokoi, S., Tamaki, S., Yano, M. & Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722 (2003).
Itoh, H., Nonoue, Y., Yano, M. & Izawa, T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nature Genetics 42, 635–U115 (2010).
Izawa, T. et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 23, 1741–55 (2011).
Putterill, J., Robson, F., Lee, K., Simon, R. & Coupland, G. The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857 (1995).
Robson, F. et al. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. The Plant Journal 28, 619–631 (2001).
Strayer, C. et al. Cloning of the Arabidopsis Clock Gene TOC1, an Autoregulatory Response Regulator Homolog. Science 289, 768–771 (2000).
Cockram, J. et al. Genome Dynamics Explain the Evolution of Flowering Time CCT Domain Gene Families in the Poaceae. PLoS ONE 7, e45307 (2012).
Satbhai, S. B. et al. Pseudo-Response Regulator (PRR) Homologues of the Moss Physcomitrella patens: Insights into the Evolution of the PRR Family in Land Plants. DNA Research 18, 39–52 (2011).
Xue, W. et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature genetics 40, 761–767 (2008).
Kim, S.-K. et al. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 228, 355–365 (2008).
Lee, Y.-S. et al. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. The Plant Journal 63, 18–30 (2010).
Wu, W. et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proceedings of the National Academy of Sciences 110, 2775–2780 (2013).
Yan, W. et al. Natural variation in Ghd7. 1 plays an important role in grain yield and adaptation in rice. Cell Research 23, 969–971 (2013).
Campoli, C., Drosse, B., Searle, I., Coupland, G. & von Korff, M. Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. The Plant Journal 69, 868–880 (2012).
Kikuchi, R., Kawahigashi, H., Oshima, M., Ando, T. & Handa, H. The differential expression of HvCO9, a member of the CONSTANS-like gene family, contributes to the control of flowering under short-day conditions in barley. Journal of Experimental Botany 63, 773–784 (2012).
Beales, J., Turner, A., Griffiths, S., Snape, J. & Laurie, D. A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theoretical and Applied Genetics 115, 721–733 (2007).
Murphy, R. L. et al. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proceedings of the National Academy of Sciences 108, 16469–16474 (2011).
Turner, A., Beales, J., Faure, S., Dunford, R. P. & Laurie, D. A. The Pseudo-Response Regulator Ppd-H1 Provides Adaptation to Photoperiod in Barley. Science 310, 1031–1034 (2005).
Yan, L. et al. The Wheat VRN2 Gene Is a Flowering Repressor Down-Regulated by Vernalization. Science 303, 1640–1644 (2004).
Salvi, S. et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad Sci U S A 104, 11376–81 (2007).
Wen, W. et al. Population structure and association mapping on chromosome 7 using a diverse panel of Chinese germplasm of rice (Oryza sativa L.). Theoretical and applied genetics 119, 459–470 (2009).
Huang, X. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nature genetics 42, 961–967 (2010).
Li, Q. et al. Cloning and characterization of a putative GS3 ortholog involved in maize kernel development. Theoretical and Applied Genetics 120, 753–763 (2010).
Liu, S. et al. Genome-Wide Analysis of ZmDREB Genes and Their Association with Natural Variation in Drought Tolerance at Seedling Stage of Zea mays L. PLoS genetics. 9, e1003790 (2013).
Chen, H., Shen, G., Wang, L. & Xing, Y. Sequence Evolution Analysis of CCT Domain Gene Family in Rice, Arabidopsis, Maize and Sorghum. JOURNAL OF HUAZHONG AGRICULTURAL UNIVERSITY 29, 669–676 (2010).
Gao, H. et al. Ehd4 Encodes a Novel and Oryza-Genus-Specific Regulator of Photoperiodic Flowering in Rice. PLoS genetics 9, e1003281 (2013).
Matsubara, K. et al. Novel QTLs for photoperiodic flowering revealed by using reciprocal backcross inbred lines from crosses between japonica rice cultivars. Theoretical and Applied Genetics 117, 935–945 (2008).
Saito, H. et al. Complete loss of photoperiodic response in the rice mutant line X61 is caused by deficiency of phytochrome chromophore biosynthesis gene. Theoretical and Applied Genetics 122, 109–118 (2011).
Yamamoto, T., Lin, H., Sasaki, T. & Yano, M. Identification of heading date quantitative trait locus Hd6 and characterization of its epistatic interactions with Hd2 in rice using advanced backcross progeny. Genetics 154, 885–891 (2000).
Yan, W.-H. et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height and heading date in rice. Molecular Plant 4, 319–330 (2011).
Wei, X. J. et al. DTH8 Suppresses Flowering in Rice, Influencing Plant Height and Yield Potential Simultaneously. Plant Physiology 153, 1747–1758 (2010).
Lin, H. X., Liang, Z. W., Sasaki, T. & Yano, M. Fine mapping and characterization of quantitative trait loci Hd4 and Hd5 controlling heading date in rice. Breeding Science 53, 51–59 (2003).
Huang, X. et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501 (2012).
Morris, G. P. et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proceedings of the National Academy of Sciences 110, 453–458 (2013).
Xing, Y., Frei, U., Schejbel, B., Asp, T. & Lübberstedt, T. Nucleotide diversity and linkage disequilibrium in 11 expressed resistance candidate genes in Lolium perenne. BMC plant biology 7, 43 (2007).
Bakker, A. et al. Reduction of hippocampal hyperactivity improves cognition in amnestic mild cognitive impairment. Neuron 74, 467–474 (2012).
McKay, S. D. et al. Whole genome linkage disequilibrium maps in cattle. BMC Genetics 8, 74 (2007).
Yan, J. et al. Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS One 4, e8451 (2009).
Fritsche, S. et al. A candidate gene-based association study of tocopherol content and composition in rapeseed (Brassica napus). Frontiers in Plant Science 3, 129 (2012).
Hassidim, M., Harir, Y., Yakir, E., Kron, I. & Green, R. M. Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 230, 481–491 (2009).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Zhao, H. et al. RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Res (2014).
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome research 12, 656–664 (2002).
Schuler, G. D. Sequence mapping by electronic PCR. Genome research 7, 541–550 (1997).
Ni, J. et al. Gramene QTL database: development, content and applications. Database: the journal of biological databases and curation 2009, bap005 (2009).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular biology and evolution 24, 1596–1599 (2007).
Librado, P. & Rozas, J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Bradbury, P. J. et al. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635 (2007).
Xu, H. et al. Genetic effects of polymorphisms in candidate genes and the QTL region on chicken age at first egg. BMC Genet 12, 33 (2011).
Lin, Y. & Zhang, Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant cell reports 23, 540–547 (2005).
Zhou, Y. et al. BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). The Plant Journal 57, 446–462 (2009).
Acknowledgements
This work was supported by grants from the National Special Program for Research of Transgenic Plant of China (2011ZX08009-001-002), the National Key Program on Basic Research and Development (2010CB125901) and the National Natural Science Foundation of China (91335201).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: Y.X. Performed the experiments: L.Z., Q.L., H.D., Q.H., L.L. and G.L. Analyzed the data: L.Z., C.T., W.Y., H.Z. and W.X. Wrote the paper: Y.X. and L.Z.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
SUPPORT
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Zhang, L., Li, Q., Dong, H. et al. Three CCT domain-containing genes were identified to regulate heading date by candidate gene-based association mapping and transformation in rice. Sci Rep 5, 7663 (2015). https://doi.org/10.1038/srep07663
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07663
This article is cited by
-
Identification of candidate genes and favourable haplotypes for yield traits in rice based on a genome-wide association study
Euphytica (2023)
-
Structural and functional analysis of CCT family genes in pigeonpea
Molecular Biology Reports (2022)
-
Meta-QTL and ortho-MQTL analyses identified genomic regions controlling rice yield, yield-related traits and root architecture under water deficit conditions
Scientific Reports (2021)
-
Opposite response of maize ZmCCT to photoperiod due to transposon jumping
Theoretical and Applied Genetics (2021)
-
Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max)
Planta (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.