Abstract
Protein fibrillogenesis is a universal tool of nano-to-micro scale construction supporting different forms of biological function. Its exploitable potential in nanoscience and technology is substantial, but the direct observation of homogeneous fibre growth able to underpin a kinetic-based rationale for building customized nanostructures in situ is lacking. Here we introduce a kinetic model of de novo protein fibrillogenesis which we imaged at the nanoscale and in real time, filmed. The model helped to reveal that, in contrast to heterogeneous amyloid assemblies, homogeneous protein recruitment is principally characterized by uniform rates of cooperative growth at both ends of growing fibers, bi-directional growth, with lateral growth arrested at a post-seeding stage. The model provides a foundation for in situ engineering of sequence-prescribed fibrous architectures.
Similar content being viewed by others
Introduction
The wealth of information protein fibers support ranges from cell locomotion1,2,3 and proliferation4,5 to neurodegeneartion6 and viral assembly7. Despite such functional diversity all protein fibers are near-crystalline assemblies exhibiting hierarchical growth8, which is believed to be polar9 and, in some cases, bidirectional10,11. Traditional techniques, such as electron and fluorescence microscopy, partly support this conjecture by providing temporal image sequences that link different points of fiber formation9,10,11,12. Notable progress has been made in the understanding of type I collagen fibrillogenesis, for which correlative measurements using electron and dark-field light microscopy revealed a unidirectional, the C-to-N terminal direction, mode of assembly13. However, direct evidence for protein fibrillogenesis from the monomer up, which would enable a kinetic-based rationale for engineering novel fibrous architectures in situ, is lacking. This can be attributed to two main reasons.
Firstly, real-time imaging of a biomolecular process has to be performed under ambient conditions in aqueous media, which excludes the use of electron microscopy. Optical microscopy is well suited for imaging dynamic processes, but is limited in spatial resolution, which has prompted the development of super-resolution techniques14,15. In particular, structured illumination microscopy (SIM) improves spatial resolution beyond the diffraction limit by illuminating the sample with patterned light to allow detection of normally unobservable high spatial frequency information. In contrast to other super-resolution techniques, SIM is capable of high imaging speeds, up to video rates and does not place additional constraints on fluorophore properties16.
Secondly, fibrillogenesis is a cooperative process programmed in the folding of monomeric units17. These units are not necessarily limited to individual polypeptide chains and may incorporate post-translational modifications that support a specific function without being directly involved in the assembly, or can comprise several populations of low oligomers, with each having different growth kinetics. An auxiliary means, additional to fiber formation, can also be employed to maintain morphological uniformity by arresting thickening18 or promote polygonal networks by seeding promiscuous interactions19. All these introduce structural polymorphism into the assembly. As a consequence, a reductionist approach is required to distinguish folding-assembly pathways from other events implicated in fibrillogenesis. In this regard, a more efficient strategy would be to replicate fiber formation from the monomer up rather than focus on resolving structural differences. De novo sequences encoding simplified folding motifs provide an excellent source of self-assembling monomers17.
With this in mind, we introduce a fibrillogenesis imaging model (FiM) – an archetypal helical assembly imaged in real time.
Results
Fibrillogenesis imaging model – design
The model is based on a de novo polypeptide sequence, KLAALKQKLAALKKELAALEQELAALEQ, designed to adopt an α-helical coiled-coil conformation – a ubiquitous structural element of α-helical fibers. Coiled coils are rope-like bundles of two or more helices interdigitated with a slight left-handed super-helical twist, which is set up by an amphipathic heptad repeat of hydrophobic (H) and polar (P) residues in their sequences, (HPPHPPP)n – usually designated abcdefg, where a and d cluster into contiguous hydrophobic interfaces20.
Native α-helical filaments adapt coiled-coil monomers by tuning their sequences into particular oligomerisation states, which alone, however, cannot set up fibrillogenesis20. Therefore, different propagation topologies and mechanisms are employed to specify the assembly, with two main principles considered in synergy.
The first principle derives from the intrinsic property of complementarity which holds true for all self-oligomerising elements. Co-assembling sequences are sufficient for controlled oligomerisation21, but may not be for directed assembly – a shortcoming which in different fibrillogenesis systems is solved by placing complementary sequences axially out of register22. For example, α-helical intermediate filaments assemble in abrupt modes23, β−structured amyloid fibrils can grow from staggered or out-of-register β−sheets24 and tropocollagen molecules assemble via side-by-side staggering11.
The second principle concerns the coiled-coil structure itself. To ensure complementary sequence offsets, engineered coiled coils are arranged to avoid polar residues at a and d sites, while relying exclusively on charge-charge interactions at g and e sites of successive heptads (g–e′ interactions)22. Thus, a staggered assembly of charged overhangs leads to extended coiled coils and fibers.
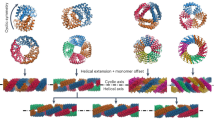
Based on this recognition, two generic heptads, cationic (KLAALKQ) and anionic (ELAALEQ), were paired sequentially into cationic and anionic halves of the designed sequence to provide oppositely charged two-heptad overhangs (Fig. 1).
FiM design.
Schematic representations of (a) the linear FiM sequence forming a homo-trimeric parallel coiled coil with two-heptad cationic and anionic overhangs, (b) a staggered assembly of the block, with continuous helical strands highlighted in black and grey (upper), with ribbon diagram of a proposed sticky-ended trimer (lower, PDB 3L4F entry rendered with PyMol) and (c) the sequence configured into coiled-coil helical wheels. Cationic and anionic heptads and residues are shown in blue and red, respectively. Double-headed arrows indicate electrostatic interactions between lysines and glutamates. Only one of the two combinations of interactions, highlighted by bold and dashed arrows, can be formed within one coiled coil (see also Supporting Information).
End-point assembly and folding
Consistent with the design rationale (see Methods), atomic force microscopy (AFM) revealed abundant homogenous fibers at micromolar chain concentrations (Fig. 2).
End-point FiM assembly.
(a) Atomic force micrographs of FiM fibers assembled overnight in 10 mM MOPS, at pH 7.4, 20°C and length histograms measured over 100 fibers (bar sizes are 200 nm). (b) Representative (upper) topography image of two individual fibers with cross-section analysis (lower) along the highlighted line.
The structures were relatively uniform in length and width, 2.7 ± 0.3 μm. Given that the peptide helix spans 4 nm and a coiled-coil diameter is approximately 2 nm these values may account for a tight lateral packing of 100 contiguous coiled-coil blocks, each comprising up to 750 super-helical units with at least four engaged electrostatic pairs per super-helix, which should render the assembly appreciably helical. Indeed, circular dichroism (CD) spectroscopy showed complete α-helix formation (100%; 100([Θ]222 + 3000)/33 000)25 (Fig. S1). As expected for helical assemblies, the peptide folded cooperatively, with thermal unfolding curves being nearly sigmoidal and CD spectra recorded before and after thermal denaturation giving similar helical contents suggesting reversibility of unfolding (Fig. S1).
Consistent with its partial destabilization the assembly was dominated by a transition midpoint (Tm) of 26°C, which, given that Tm values for four electrostatically complete heptads (eight electrostatic pairs) are in a typical range of 40–60°C8,26, was in good agreement with Tm drops of 20–30°C as a result of incomplete or disrupted electrostatic networks26. Low transition temperatures combined with cooperativity in folding and fiber uniformity imply that the assembly matures rapidly and should be complete within a few hours. This is essential for continuous imaging at room temperature and suggests that fibrillogenesis should proceed within the first hours from lower oligomers27,28. What remains unclear is whether such oligomers present one growth phase, longitudinal or lateral, both phases that are equally favored for propagation or both phases with one reaching earlier maturation. The latter scenario appears to be characteristic of amyloid fibrils and collagen fibers10,11 and is supported by fiber tapering – the effect which is also evident from the AFM images of FiM fibers. This is likely to indicate a simultaneous growth of both ends, albeit at possibly different rates (Fig. 2). Importantly, tapering emphasizes the uniformity of widths along the main fiber body, which would be consistent with an earlier maturation of a lateral growth phase followed by longitudinal expansion in both directions. Recently, amyloid-like fibrils were shown to undergo bidirectional elongation starting from seed oligomers29. However, large heterogeneities were observed in growth rates, from no growth to 10 times the original seed size, which are deemed due to the structural polymorphism of amyloid assemblies29. This is in drastic contrast to FiM designed to provide statistically significant fiber densities for the analysis of homogeneous growth kinetics.
Real-time assembly and growth kinetics
Optical microscopy images confirmed uniform FiM growth and revealed a dense carpet of individually distinguishable fibers (Fig. 3a). Time-lapse imaging performed for FiM mixed with FiM-Alexa488 (1:10−3 ratios at micromolar chain concentrations) at 30-sec intervals, from sample preparation till maturation, showed that fibers grew bi-directionally to full (~2.5 μm) lengths within 3–5 hours (Fig. 3a and Video 1).
Real-time FiM assembly.
(a) Widefield and SIM micrographs of fibers assembled from FiM mixed with FiM-Alexa488 (1:10−3 molar ratios). Green fluorescence is shown only in green-filtered squares to emphasize contrast against the fluorescent background. (b) (right) Growth kinetics of individual fibers highlighted in the micrograph (left). Bold lines are 4th order polynomical fits to the raw data. Inset highlights growth for fiber 2 at both ends, tip growth; using two different colours (see also Fig. S2–S4).
To gain a better insight into growth kinetics, individual fibers in images were segmented from the background and tracked for further analysis (Fig. 3b, S2 and Video 2). The segmentation of fibers in Fig. 3a indicates a fiber confluency (fraction of substrate area covered by fibers) of ~40% of the total image area (Fig. S3). Starting immediately after peptide preparation (0% confluency) there was an initial period of relatively rapid growth (~20 nm per min), after which the growth rate gradually settles at a plateau of 40% confluency that remained stable overnight (Fig. 3b, S3). Such behaviour indicates equilibrium between peptide monomers in solution and in the assembly, at which point the assembly is essentially mature. Maturation proved to be highly cooperative and characteristic of individually homogeneous growth (Fig. 4a and Video 3), including cases of proximity-driven fusions between fibers, which tended to grow to full lengths before joining up (Fig. 4a and Video 4).
Real-time individual FiM assembly.
(a) Single (left) and fusion (right) fiber assemblies imaged over 240 min with individual images separated at 30-min intervals. (left) The bright round feature to the left of the center is a fluorescent aggregate that can be used for image registration. Scale bars are 2 μm. (b) Proposed bottom-up three-phase assembly model for homogeneous protein fibrillogenesis comprising I-phase – seeding phase of monomeric units, initiation; S-phase – laterally equilibrated assemblies, stabilization and M-phase – longitudinally equilibrated assemblies, maturation. Key: blue rings denote equilibrated (arrested) growth and arrows denote continuous growth. Cylinders with a coiled-coil trimer correspond to a monomeric unit.
Discussion
Collectively these measurements prompt two conclusions, both being in strong support of one-mode elongation independent of immediate oligomer environment, which supports continuous, albeit uni-polar, growth of collagen type I fibrils13 and both being in contrast to current models of heterogeneous amyloid and amyloid-like assemblies27,28,29,30,31.
Firstly, after the formation of seed assemblies, within the first hour of imaging, no width increases were apparent implying that at that point radial assembly was arrested or stabilized, with both longitudinal and radial growths being at comparable rates (Fig. 4a and Videos). This observation suggests that an initiation or seeding phase precedes a first growth phase, stabilization, whereupon size increases are limited exclusively to length increases, a second growth phase – maturation, which eventually reaches elongation plateau (Fig. 3b). Secondly, growth rates at both ends of individual fibers, tip growth, proved to be nearly identical (Fig. 3b, S4 and Video 2), which indicates that same monomeric units recruit at both ends with similar affinities32. This stands for a bi-directional mode of assembly best described by a three-phase growth model as follows: (i) cooperatively folded monomeric units (coiled-coil trimers or higher oligomers) assemble into seed structures, which upon reaching a threshold in radial growth (ii) stabilize into equilibrated assemblies that (iii) continue growing longitudinally at thermodynamically favorable rates (Fig. 4b)33.
Irrespective of biological function and origin protein fibrillogenesis is spontaneous, but directional, reaching a point of maturation, which is accompanied by lateral adjustments of early oligomers or low fibrils. For instance, super-coiling in filamentous helices can seed extended super-helices which then spontaneously associate in a head-to-tail fashion into thicker filaments34. Conversely, twisting in collagen fibrils tends to limit radial growth in collagen fibrillogenesis35, whereas fine tuning in coiled-coil interfaces can arrest thickening36. Growth polarity however remains the limiting factor of maturation driving fiber formation to completion.
Contiguous nanometer periodicities (D-periodicity in collagens11 or striation patterns in α-helical fibers8) as well as shape polarity (coarse and fine tapered tips in collagen fibrils)35 strongly support a unipolar mode of assembly. Analogous to the direction of polypeptide chains running from the amino end to the carboxyl end (N-to-C), a self-assembling fiber can also have N- and C ends13, with some collagens being N,N- and molecularly bipolar4,11,37. Both unipolar and bipolar fibrils can elongate via tip fusions into longer structures, in which case unipolar fibrils act as building blocks while bipolar fibrils are capping elements38. Thus, the ratio of unipolar/bipolar fibrils can determine the form, composition and size of resulting fibers. Similarly, amyloid-like fibrils mature through the fusion of seed oligomers whose composition and size can be different and growth kinetics substantially varied29. In these cases fibrillogenesis is challenged by heterogeneity, can be compromised and in some cases inhibited.
To avoid such complications in revealing mechanistic insights into homogeneous fibrillogenesis an idealized self-assembly model is needed first. Such a model should facilitate the extraction of assembly mechanisms that are free from the constraints of structural polymorphism and has to be structurally, synthetically and temporally accessible to simplify and minimize sample preparation, which may otherwise affect the accuracy of results. Granted such a model, the assembly process may then be monitored in real time using direct live imaging, filming.
Based on these conventions, we have provided the first de novo and real-time model of homogeneous fibrillogenesis from the monomer up that is principally characterized by uniform and cooperative growth at both ends– a mechanistic rationale that has not been shown before. In this model, the transition from stabilization to maturation is independent of the continuous formation of seed structures and their fusion, which offers a kinetic-based rationale for monitoring and building engineered architectures in situ.
Methods
FiM design notes
Alanines and glutamines were used at the solvent-exposed b, c and f sites to minimize side-by-side, uncontrolled association of coiled-coil oligomers and to have a stabilizing effect on fibre thickening8, which is further supported by a lysine at a single f site. The lysine introduces an overall positive net charge and was used to enable site-specific fluorescent labelling on resin during polypeptide synthesis. Leucines were placed at a and d to favor trimers and to loosen electrostatic interactions by freeing up some g and e sites, which would otherwise support dimer formation20. Individual electrostatic interactions are thus unfixed in the same coiled-coil block and any g–e′ pair of one helical pair can be replaced by a mirror g–e′ pair of another helical pair. Unengaged g–e′ pairs at any given time make coiled-coil and fiber surfaces charged and highly hydrophilic thereby thermodynamically disfavoring further fiber thickening in aqueous media. All pair re-engagements are concomitant along the aligned sequences which renders any changes in the entire electrostatic network highly cooperative. This measure was introduced for three reasons: firstly, to shorten the assembly and maturation time at the expense of partial destabilization of the coiled coil and subsequently arresting fiber growth, which would, secondly, allow continuous imaging under equilibria conditions at room temperature that at longer time periods can be compromised by increasing fiber precipitation and bundling as well as water evaporation. Thirdly, partially destabilized coiled coils were expected to give rise to homogenous and medium-sized fibers, a few microns in length and in densities that would be sufficiently high for statistical analysis within an image frame.
Peptide synthesis
All peptides were assembled on a Liberty microwave peptide synthesizer (CEM Corp) using Fmoc-Gln(Trt)-Wang resin, standard solid-phase Fmoc/tBu protocols and HBTU/DIPEA as coupling reagents. Fmoc-Lys(Alloc)-OH was used for the orthogonal coupling of Alexa-488 to the peptide (Figure S5). After cleavage and deprotection (95% TFA, 2.5% TIS, 2.5% water) peptides were purified using RP-HPLC and their purities were confirmed by analytical RP-HPLC and MALDI-ToF (Figure S6). MS [M + H]+: FiM – m/z 3033.65 (calc), 3033.7 (observed); FiM-Alexa488– m/z 3555.65 (calc), 3557.3 (observed). [M + Na]+ ions were also observed.
High performance liquid chromatography
Analytical and semi-preparative gradient RP-HPLC was performed on a JASCO HPLC system using Vydac C18 analytical (5 μm) and semi-preparative (5 μm) columns. Both analytical and semi-preparative runs used a 10-70% B gradient over 50 min at 1 mL/min and 4.7 mL/min respectively with detection at 230 and 220 nm. Buffer A – 5% and buffer B – 95% aqueous CH3CN, 0.1% TFA.
FiM assembly
Aqueous peptide solutions (200 μL) containing FiM (300 μM) mixtures with FiM-Alexa488 at the 1:10−3 molar ratio were prepared in filtered (0.22 μm) 10 mM MOPS, pH 7.4 and used for CD, AFM and optical microscopy studies. The ratio was experimentally optimized to allow for direct fluorescence imaging while minimizing interference from background fluorescence.
Circular dichroism spectroscopy
Circular dichroism spectroscopy was performed on a JASCO J-810 spectropolarimeter fitted with a Peltier temperature controller. All measurements were taken in ellipticities in mdeg and after baseline correction converted to mean residue ellipticities (MRE, deg cm2 dmol-res−1) by normalizing for the concentration of peptide bonds and cuvette path length. Contributions to the CD from FiM-Alexa488 were neglected.
Atomic force microscopy
For AFM imaging of a clean silicon wafer immerse in a 300 μM solution of peptide in 10 mM MOPS (pH 7.4) and incubated overnight at room temperature was analyzed. Topographic, amplitude and phase AFM images were recorded using tapping mode AFM on a Cypher Instrument (Asylum Research). All the AFM images were flattened with a first order line-wise correction fit. AFM tips used were super-sharp silicon probes (Nanosensors; resonant frequency ~330 kHz, tip radius of curvature, 5 nm, force constant 42 N/m). Images were processed using proprietary software, SPIP 6.0.2.
Fluorescence microscopy
Optical imaging of fibers assembled from FiM mixed with FiM-Alexa488 peptides was performed using a custom-built microscope system developed around a commercially available inverted microscope body (IX71, Olympus). Sinusoidal excitation patterns for structured illumination microscopy (SIM) were created using a ferroelectric liquid crystal on silicon spatial light modulator (SXGA-3DM, Fourth Dimension Displays) (SLM) configured to display a series of binary phase gratings15. The SLM was illuminated with collimated light from a 488 nm laser (Sapphire LP, Coherent Inc.) and imaged onto the sample through the objective lens. A spatial filter in the Fourier plane of the SLM passed only the positive and negative first diffracted orders which were imaged onto the pupil plane of the objective lens. All images were acquired using a scientific CMOS camera (Orca Flash 4.0 v1, Hamamatsu Photonics) with the global exposure period of the camera's rolling shutter synchronized with the display of gratings on the SLM. A closed loop focus stabilization system (CRISP, Applied Scientific Instrumentation) was fitted to the microscope to minimize focal drift during time-lapse imaging. SIM images of the resulting Alexa488-labelled fibers were acquired using a 60x/1.3 objective lens (UPLSAPO 60XS, Olympus). Each SIM image was reconstructed from nine raw images of the sample acquired under with different sinusoidal illumination patterns (three orientations and three phase steps per orientation) with images reconstructed as described16. Time-lapse total internal reflection fluorescence (TIRF) images of the self-assembling Alexa488-labelled fibers were acquired using a 100X/1.49 objective lens (UAPON 100XOTIRF, Olympus). For TIRF imaging the period of the grating displayed on the SLM was set to ensure that the first diffracted orders were incident inside the TIRF ring of the objective lens pupil. In principle, the nine raw images acquired under sinusoidal TIRF illumination could be processed to reconstruct a super-resolution image as in the widefield case16. In practice, owing to the relatively low signal-to-noise ratio of the images, effective reconstruction of reliable super-resolution TIRF images was not possible. For this reason the nine raw images acquired under sinusoidal illumination were averaged to give a single diffraction-limited TIRF image with improved illumination uniformity and signal-to-noise ratio.
Segmentation and tracking of individual fibers
Drift induced lateral offsets between images in each time-lapse image sequence were removed using intensity-based registration, performed with bright aggregated peptide clusters used as fiducial markers. The intensity of each image in the sequence was then normalized to the brightest pixel and a morphological top hat filter was applied to increase contrast. Individual peptide fibers were segmented and tracked through the time-lapse sequence using open active contours implemented in the ImageJ plugin JFilament39. Foreground and background pixel values were defined individually for each fiber tracked in the sequence. All other contour parameters were empirically optimized once and assigned the same values for all fibers in the sequence.
References
Grosse, R. & Vartiainen, M. K. To be or not to be assembled: progressing into nuclear actin filaments. Nat. Rev. Mol. Cell. Biol. 14, 693–697 (2013).
Abu Shah, E. & Keren, K. Mechanical forces and feedbacks in cell motility. Curr. Opin. Cell. Biol. 25, 550–557 (2013).
Chung, B. M., Rott, J. D. & Coulombe, P. A. Networking galore: intermediate filaments and cell migration. Curr. Opin. Cell. Biol. 25, 600–612 (2013).
Mienaltowski, M. J. & Birk, D. Structure, physiology and biochemistry of collagens. Adv. Exp. Med. Biol. 802, 5–29 (2014).
Wen, Q. & Janmey, P. A. Effects of non-linearity on cell-ECM interactions. Exp. Cell. Res. 319, 2481–2489 (2013).
Ellis, R. J. Protein misassembly: macromolecular crowding and molecular chaperones. Adv. Exp. Med. Biol. 594, 1–13 (2007).
Marvin, D. A., Symmons, M. F. & Straus, S. K. Structure and assembly of filamentous bacteriophages. Prog. Biophys. Mol. Biol. 114, 80–122 (2014).
Papapostolou, D. et al. Engineering nanoscale order into a designed protein fiber. Proc. Natl. Acad. Sci. USA 104, 10853–10858 (2007).
Smith, A. M. et al. Polar assembly in a designed protein fiber. Angew. Chem. Int. Ed Engl. 44, 325–328 (2004).
Scheibel, T., Kowal, A. S., Bloom, J. D. & Lindquist, S. L. Bidirectional amyloid fiber growth for a yeast prion determinant. Curr. Biol. 11, 366–369 (2000).
Starborg, T., Lu, Y., Meadows, R. S., Kadler, K. E. & Holmes, D. F. Electron microscopy in cell-matrix research. Methods 45, 53–64 (2008).
Esbjörner, E. K. et al. Direct observations of amyloid β self-assembly in live cells provide insights into differences in the kinetics of Aβ(1-40) and Aβ(1–42) aggregation. Chem. Biol. 21, 732–742 (2014).
Kadler, K. E., Hojima, Y. & Prockop, D. J. Collagen fibrils in vitro grow from pointed tips in the C- to N-terminal direction. Biochem J. 268, 339–343 (1990).
O'Holleran, K. & Shaw, M. J. Optimized approaches for optical sectioning and resolution enhancement in structured illumination microscopy. Biomed. Opt. Express 5, 2580–2590 (2014).
Axelrod, D. Total internal reflection fluorescence microscopy in cell biology. Traffic 2, 64–774 (2001).
Kner, P., Chun, B. B., Griffis, E. R., Winoto, L. & Gustafsson, M. G. L. Super-resolution video microscopy of live cells by structured illumination. Nat. Methods 5, 339–342 (2009).
Woolfson, D. N. & Ryadnov, M. G. Peptide-based fibrous biomaterials: Some things old, new and borrowed. Curr. Opin. Chem. Biol. 10, 559–567 (2006).
Wagner, D. E. et al. Toward the development of peptide nanofilaments and nanoropes as smart materials. Proc. Natl. Acad. Sci. USA 102, 12656–12661 (2005).
Bella, A., Ray, S., Shaw, M. & Ryadnov, M. G. Arbitrary self-assembly of peptide extracellular microscopic matrices. Angew. Chem. Int. Ed. 51, 428–431 (2012).
Parry, D. A., Fraser, R. D. & Squire, J. M. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J. Struct. Biol. 163, 258–269 (2008).
Harbury, P. B., Zhang, T., Kim, P. S. & Alber, T. A switch between two-, three- and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262, 1401–1407 (1993).
Ryadnov, M. G., Bella, A., Timson, S. & Woolfson, D. N. Modular design of peptide fibrillar nano- to microstructures. J. Am. Chem. Soc. 131, 13240–1324 (2009).
Chernyatina, A., Nicolet, S., Aebi, U., Herrmann, H. & Strelkov, S. V. Atomic structure of the vimentin central α-helical domain and its implications for intermediate filament assembly. Proc. Natl. Acad. Sci. USA 109, 13620–13625 (2012).
Liu, C., et al. Out-of-register β-sheets suggest a pathway to toxic amyloid aggregates. Proc. Natl. Acad. Sci. USA. 109, 20913–20918 (2012).
Morrisett, J. D., Jackson, R. L. & Gotto, A. M. Lipid-protein interactions in the plasma lipoproteins. Biochim. Biophys. Acta 472, 93–133 (1977).
Ciani, B. et al. Molecular basis of coiled-coil oligomerization-state specificity. Proc. Natl. Acad. Sci. USA 107, 19850–19855 (2010).
Bromley, E. H. C. et al. Assembly pathway of a designed α-helical protein fiber. Biophys. J. 98, 1668–1676 (2010).
Yokoi, H., Kinoshita, T. & Zhang, S. Dynamic reassembly of peptide RADA16 nanofiber scaffold. Proc. Natl. Acad. Sci. USA 102, 8414–8419 (2005).
Pinotsi, D. et al. Direct observation of heterogeneous amyloid fibril growth kinetics via two-color super-resolution microscopy. Nano Lett. 14, 339–345 (2014).
Kellermayer, M. S., Karsai, A., Benke, M., Soós, K. & Penke, B. Stepwise dynamics of epitaxially growing single amyloid fibrils. Proc. Natl. Acad. Sci. USA 105, 141–144 (2008).
Ban, T. et al. Direct observation of Abeta amyloid fibril growth and inhibition. J Mol. Biol. 344, 757–67 (2005).
Ryadnov, M. G. & Woolfson, D. N. MaP peptides: programming the self-assembly of peptide-based mesoscopic matrices. J. Am. Chem. Soc. 127, 12407–12415 (2005).
Williams, R. J. et al. Enzyme-assisted self-assembly under thermodynamic control. Nat. Nanotechnol. 4, 19–24. (2009).
Herrmann, H. & Aebi, U. Intermediate filaments: molecular structure, assembly mechanism and integration into functionally distinct intracellular scaffolds. Annu Rev Biochem. 73, 749–789 (2004).
Holmes, D. F., Chapman, J. A., Prockop, D. J. & Kadler, K. E. Growing tips of type I collagen fibrils formed in vitro are near-paraboloidal in shape, implying a reciprocal relationship between accretion and diameter. Proc. Natl. Acad. Sci. USA 89, 9855–9859 (1992).
De Santis, E., Faruqui, N., Noble, J. E. & Ryadnov, M. G. Exploitable length correlations in peptide nanofibres. Nanoscale 6, 11425–11430 (2014).
Thurmond, F. A. & Trotter, J. A. Native collagen fibrils from echinoderms are molecularly bipolar. J Mol Biol. 235, 73–79 (1994).
Kadler, K. E., Holmes, D. F., Trotter, J. A. & Chapman, J. A. Collagen fibril formation. Biochem. J, 316, 1–11 (1996).
Smith, M. B. et al. Segmentation and tracking of cytoskeletal filaments using open active contours. Cytoskeleton 67, 693–705 (2010).
Acknowledgements
We acknowledge funding from the UK's Department of Business, Innovation and Skills and the Strategic Research Programme of the National Physical Laboratory.
Author information
Authors and Affiliations
Contributions
A.B. and M.S. contributed equally to this work. All authors designed experiments and analysed the data. A.B, M.S. and S.R. performed the experiments. M.G.R wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Video 1
Supplementary Information
Video 2
Supplementary Information
Video 3
Supplementary Information
Video 4
Supplementary Information
Supporting Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Bella, A., Shaw, M., Ray, S. et al. Filming protein fibrillogenesis in real time. Sci Rep 4, 7529 (2014). https://doi.org/10.1038/srep07529
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07529
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.