Abstract
We provide direct evidence that Bin/Amphiphysin/Rvs (BAR) family members bend the steady state membrane architecture of organelles in intact cells. In response to inducible BAR molecular actuators, organelles exhibit distinct changes to the orientation and degree of their membrane curvature. This rapidly inducible system may offer a mechanism by which to better understand the structure-function relationship of intracellular organelles.
Similar content being viewed by others
Introduction
Cellular organelles possess unique morphological characteristics that underlie their function. Defects in the organization of their membrane structure have devastating effects on physiological processes that can directly contribute to diseases1. Despite this significance, the mechanism by which organelles are shaped three-dimensionally is not well understood. Experimental techniques to manipulate intracellular membrane morphology should greatly enhance our understanding of the structure-function relationship of intracellular membranes. One strategy would be to engineer a native membrane-deformation process. Proteins including Bin/Amphiphysin/Rvs (BAR) family members sense and/or induce membrane deformation using a variety of mechanisms, including asymmetric protein distribution, crowding, scaffolding and hydrophobic insertion1,2. BARs contain conserved domains that dimerize to shape a boomerang of varying arc length2,3,4,5,6 which are categorized in three main subfamilies: the classic BAR (also called N-terminal amphipathic helix-containing BAR, N-BAR), Fes/CIP4 homology BAR (F-BAR) and inverse BAR (I-BAR)3. These domains bind through electrostatic interactions to negatively charged lipid membranes and mediate membrane curvature through scaffolding and/or amphipathic helix insertion along lipid bilayers, independent of biochemical signaling mechanisms2,3,4. However, direct evidence for BAR-induced intracellular membrane deformation is currently lacking. This is mainly due to the difficulty of performing experiments in vivo with rapid onset of BAR accumulation. Bending assays with BAR domains to date have typically been conducted in vitro using purified BARs added to lipid vesicles, demonstrating rapid tubulation of the membrane upon addition of these proteins3,7,8. While lipid vesicles provide a convenient, minimal experimental environment, they do not faithfully represent biological membranes due to a lack of diverse constituent lipids, membrane-interacting proteins, or cortical actin. In contrast, overexpression of BAR proteins in cell culture offers an understanding of how these domains behave in cells, yet the slow accumulation of BAR domains at membranes over time may give rise to secondary effects due to other unaccounted biological contributions. In order to complement these limitations and to visualize membrane bending in situ in real time, we employed a chemically inducible dimerization (CID) technique that allows for the second-by-second recruitment of a protein of interest to a specific organelle in intact live cells. In this study, we demonstrate that the CID-engineered BARs can rapidly deform plasma membrane as well as intracellular membranes in living cells.
Results
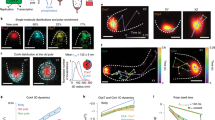
The CID technique relies on two proteins, FK506 binding protein (FKBP) and FKBP-rapamycin binding domain (FRB), binding to one another upon introduction of a chemical dimerizer such as rapamycin9. Using this system, we have previously anchored FRB to the cytosolic face of various individual organelles to which FKBP, now fused to a protein-of-interest, was rapidly recruited10. In the present study, we constructed a new series of FKBP fusion proteins that incorporate BAR domains with different intrinsic curvature properties (left panels, Fig. 1). These BAR domains are taken from the human Amphiphysin 1 (N-BAR), formin binding protein 17 (F-BAR), Missing-in-Metastasis (I-BAR) as well as their respective mutants impaired in curvature-inducing activity: amphipathic helix deletion mutant (ΔN-BAR) or electrostatic mutants (F-BAR-QQ and I-BAR(12,15)). We then transfected COS-7 fibroblast-like cells with two sets of constructs: one from the FKBP-BAR fusion library and one from the FRB-anchor library that constitutively localizes either to plasma membrane, mitochondria, endoplasmic reticulum (ER), or Golgi (Supplementary Table S1). Organelle morphology was visualized under confocal and epifluorescence microscopes as shown by organelle markers (Figs. 1–2) and recruitable BARs (Supplementary Figs. S1–2), while the cells were simultaneously treated with rapamycin. Any material observations consistent with strict criteria (Supplementary Table S2) were quantified and recorded as the percentage of cells displaying the phenotype (Table 1). We describe the results categorized by BAR domain subtype.
Recruitment of BAR domains to intracellular locations triggers distinctive morphological changes as shown by membrane markers.
Paired with Supplementary Fig. S1. Representative images of COS-7 cells expressing an organelle anchor unit and recruitable unit (fluorescent protein-FKBP-BAR and fluorescent protein-FRB-anchor unit). For simplicity, recruitable BARs are listed as BAR cartoons to the left of images and anchor units are described above images as their sites of localization. Cartoons of BAR proteins: human Amphiphysin 1 N-BAR, PDB: 4ATM; human FBP17 F-BAR, PDB: 2EFP; human MIM I-BAR, PDB: 2D1L. Note that the cartoon from the crystal structure of Amphiphysin 1 does not show the N-terminal amphipathic helix. Cells were imaged at 15-second intervals for at least 2 minutes and 30 seconds prior to addition of 100 nM rapamycin for a total of 15 minutes. At least 3 independent trials of more than 30 cells were analyzed. Images show the FRB-anchored at organelles before and after rapamycin addition. Red arrowheads highlight morphological changes of membranes. Scale bars indicate 2 μm.
Recruitment of BAR-mutant and FKBP-controls to intracellular locations as shown by membrane markers.
Paired with Supplementary Fig. S2. Representative images of COS-7 cells expressing an organelle anchor unit and recruitable unit (fluorescent protein-FKBP-mutant BAR or fluorescent protein-FKBP and fluorescent protein-FRB-anchor unit). For simplicity, recruitable BAR-mutants are listed as cartoons and fluorescent protein-FKBP is designated as “FKBP only” to the left of images. Anchor units are described above images as their sites of localization. Cells were imaged at 15-second intervals for at least 2 minutes prior to addition of 100 nM rapamycin for a total of 15 minutes. At least 3 independent trials of more than 30 cells were analyzed. Images show the FRB-anchored organelles of BAR-mutants and FKBP- control conditions before and after rapamycin addition. Scale bars indicate 2 μm.
N-BAR
Rapid recruitment of N-BAR promoted the appearance of tubulations at the plasma membrane ranging from 2.1 to 33.6 μm in length and the percentage of cells displaying this phenotype was 36.0 +/− 13.1%. As expected, ΔN-BAR mutant and FKBP negative controls did not result in plasma membrane tubulation. To further investigate the observation at an ultra-structural level, we performed cryo-immunogold electron microscopy where an antibody against a fluorescent protein tagging FKBP-BAR was used. Both N-BAR and ΔN-BAR mutant electron micrographs clearly indicated the immunogold labeling at the plasma membrane (Fig. 3a). Although N-BAR and ΔN-BAR showed similar degrees of labeling of the surface plasma membrane, N-BAR showed greater labeling of the intracellular tubules compared to the ΔN-BAR control (Fig. 3b). It should be noted that these micrographs did not capture the plasma membrane invaginating to form tubular structures.
Electron micrographs of the plasma membrane region.
Anti-GFP immunogold labeling of COS-7 cells expressing Lyn-FRB and YFP-FKBP-N-BAR or Lyn-FRB and GFP-FKBP-ΔN-BAR. (a) Immunogold labeling of YFP-FKBP-N-BAR shows localization at the plasma membrane and internal membrane. These labeled internal membrane tubules may reflect the tubules we observe under light microscopy. Less immunogold labeling of internal membrane tubules occurs in the GFP-FKBP-ΔN-BAR negative control sample. The p value calculated from unpaired, two-tailed Student's t-test is 0.81. (b) Quantification of the ratio of gold particles at the plasma membrane surface to the overall amount of labeling is similar for YFP-FKBP-N-BAR and the negative control condition. YFP-FKBP-N-BAR scaffolds internal membrane tubules, as measured within 100 nm of the internal tubule, while the negative control has significantly less labeling along similar tubules. The p value calculated from unpaired, two-tailed Student's t-test is less than 0.0001. Scale bar indicates 500 nm.
In contrast to the plasma membrane phenotype, recruitment of N-BAR to ER gave rise to punctate structures 1–3 μm2 along the tubular network and partially to ER sheets at a frequency of 55.1 +/− 14.5%. Interestingly, ΔN-BAR did not significantly reduce the ER phenotype (36.9 +/− 11.8%). Recruitment of N-BAR to the cis- and medial- Golgi caused tubulations and protrusions of the organelle ranging in diameter from 0.2 to 1.9 μm and extended at a rate less than 0.01 μm/sec at a frequency of 47.0 +/− 2.6%. These protrusive structures emanating from the Golgi are distinct from the dynamic, naturally occurring tubules that extend from the Golgi stacks as previously described11. Recruitment of ΔN-BAR significantly reduced the frequency of the phenotype (25.0 +/− 8.3%). Finally, there was no visible structural change to mitochondria upon recruitment of N-BAR.
F-BAR
F-BAR recruitment to the ER resulted in the formation of puncta 1–3 μm2 along the ER tubular and sheet-like network at a frequency of 48.9 +/− 1.9%, a phenotype similar to that induced by N-BAR. The F-BAR mutant counterpart deficient in positively charged lipid-binding residues (F-BAR(QQ)) induced this change, albeit mildly (17.0 +/− 8.9%). No other organelles showed morphological changes in response to F-BAR recruitment.
I-BAR
I-BAR recruitment to plasma membrane and mitochondria triggered the formation of thin protrusions. At the plasma membrane, filopodia-like protrusions extended 3.1 to 5.1 μm in length at a frequency of 74.0 +/− 4.1%. In contrast, I-BAR(12,15) reduced the frequency of the phenotype (14.9 +/− 2.1%). At the mitochondria, unusual protrusions elongated at a rate of more than 0.2 μm/sec at a frequency of 60.9 +/− 5.1% as a result of I-BAR recruitment, which is statistically significant relative to I-BAR(12,15) (9.9 +/− 4.0%). Recruitment of I-BAR to the ER resulted in punctate structures 1 μm2 in size at a frequency of 100.0 +/− 0.0%, morphologically distinct from those induced by N-BAR and F-BAR. The resulting phenotype appeared similar to the fusion of ER and the plasma membrane (Supplementary Fig. S3). Recruitment of I-BAR(12,15) resulted in unique punctate structures 50 to 60 μm2 at a frequency of 79.7 +/− 5.8% and occurred largely at sheet-like ER (Fig. 2 and Supplementary Fig. S3). Recruitment of I-BAR to Golgi did not show any detectable morphological changes.
To test whether the relative expression level or percentage of overexpressed FKBP-BAR translocation to organelles influenced the frequency of the observed phenotypes, we further analyzed the condition of N-BAR induced tubulations at the plasma membrane. Specifically, we quantified the appearance of tubules at the plasma membrane following N-BAR recruitment versus the fluorescence intensity of N-BAR overexpression or percentage of FKBP-BAR translocation per unit area of the cell (Supplementary Fig. S4). Equations could not be fitted to the data to show a trend, suggesting that BAR concentration has no relationship with the resulting membrane phenotype, at least within the range of BAR expression levels in our experiments.
Lastly, we tested if molecular crowding itself drives membrane bending by recruiting FKBP alone (sans BARs) to each organelle. As a result, we did not observe any of the above changes driven by BARs (Fig. 2), suggesting that properties unique to BARs promote the observed organellar membrane phenotypes. BARs were also recruited to other organelles such as endosomes, lysosomes and inner nuclear membrane. However, no obvious structural differences emerged (data not shown). These organelles may retain sufficient structural rigidity and lipid packing so as not to be receptive to our technique12. Morphological changes may have also been missed due to the limited spatial resolution of fluorescence microscopy.
Discussion
Our results unambiguously demonstrated that BAR domains induce membrane curvature in an organelle-specific manner in live cells on a timescale of seconds. This curvature is dependent on the unique characteristics of each BAR subtype. These differential effects are intriguing in terms of how precisely this bending occurs and how the resulting morphology relates to endogenous cellular events. A number of different mechanisms are responsible for the resulting morphology of cells and organelles. Lipid packing, lipid composition, integral membrane protein localization or wedge-like insertion of peripheral membrane proteins, protein crowding, protein scaffolding and cytoskeletal-based mechanisms contribute to curvature of intracellular membranes1,2. Previous studies on BAR-induced membrane curvature performed in vitro, in vivo and in silico also demonstrated that tubule diameters vary based on factors ranging from BAR concentration, type of BAR, oligomerization status and the dielectric constant of the lipid membrane to the local environment13,14. Given the complexity in the lipid and protein composition of organelles, we thus expect that BAR recruitment to these locations would result in a variety of morphological effects. Indeed, we observed a range of phenotypes occurring at different organelles, whose frequency is not directly correlated with the fluorescence intensity of overexpressed FKBP-BARs.
Our study has indicated the characteristics of FKBP-BAR-induced phenotypes as being reminiscent of actual cellular morphologies, both typical and aberrant. At the plasma membrane, endocytosis exemplifies a cellular process of curvature generation where membrane tubulation and vesicle budding is essential1. Using the CID system, we observed membrane tubules forming upon N-BAR recruitment in both live-cell epifluorescence images and electron micrographs. In contrast, F-BAR recruitment to the plasma membrane did not result in tubulations, in contrast to previous studies in vivo and in vitro7. It is possible that the FKBP-fusion proteins alter or even diminish the curvature inducing properties of the individual BAR domains themselves. Next, in processes related to cell migration, finger-like projections such as filopodia arise from outward growing filamentous actin pushing against the membrane, to which I-BAR proteins are recruited15. Similarly, recruitment of I-BAR via the CID system to the plasma membrane resulted in filopodia-like structures. At the mitochondria, membrane sculpting proteins such as Dmn1/Drp1 regulate the dynamic network of tubules that undergo fusion and fission16. Along a similar theme, I-BAR recruitment with our system resulted in dynamic protrusions of the mitochondria. The ER is composed of a tubular and sheet-like network shaped by architectural proteins such as reticulins and atlastins17. Mutations in the reticulin family members deform the ER into punctate-like structures along the tubular network. Interestingly, CID-based recruitment of N-BAR and F-BAR produced puncta along the ER network, while I-BAR and the I-BAR(12,15) produced other distinctive punctate structures. The Golgi undergoes broad changes in morphology as a key organelle in the secretory pathway. A variety of tubules extend throughout the stacks of cis-, medial- and trans- Golgi11. Recruitment of N-BAR resulted in larger tubulations than those already occurring. It is notable that BAR recruitment to these different intracellular locations resulted in phenotypes already observed naturally under specific conditions. The rapidly recruitable BAR technique developed in the present study may thus offer a powerful method to investigate cellular events associated with the morphological changes under both physiological and pathophysiological conditions.
In summary, organelle shape has long been thought to be fundamental to cellular function. Yet, it has been difficult to study the structure-function relationships of individual organelles because structural perturbations have been primarily driven by genetic manipulations where the “architectural” roles of proteins are not easily distinguishable from their “signaling” ones. The molecular actuators introduced here perturb organelle shape on a second timescale in live cells where we can simultaneously measure biologically relevant cellular activity. Their use permits insights into how specific organelles contribute to health and disease such as neuropathies associated with altered ER morphology and neurodegenerative diseases linked to altered Golgi and mitochondrial structure.
Methods
DNA constructs and other reagents
We received the constructs encoding human Amphiphysin 1 N-BAR and human FBP17 F-BAR from Dr. Toshiki Itoh (Kobe University) and human MIM I-BAR from Dr. Pekka Lappalainen (University of Helsinki). BARs were subcloned into FKBP vectors10 between BamHI and EcoRI restriction sites. These DNA constructs include BARs encoding the N-BAR from human Amphiphysin 1 (1–248 AA), curvature-inducing deficient mutant ΔN-BAR (26–248 AA), F-BAR from human forming-binding protein (FBP17) (1–300 AA), lipid binding deficient mutant F-BAR (R113Q + K114Q), I-BAR from human missing-in-metastasis (MIM) (1–254 AA) and I-BAR (12,15) mutant (K146A, K149A, K150A, K138A, K139A). The anchor units Lyn-fluorescent protein-FRB at the plasma membrane, fluorescent protein-FRB-MoA at the mitochondria, fluorescent protein-FRB-Cb5 at the ER, or fluorescent protein-FRB-Giantin at the cis- and medial- Golgi were previously described10.
Tissue culture and transfection
COS-7 cells were cultured in DMEM (Gibco) + 10% FBS media. DNA constructs were transfected with FuGENE HD (Roche).
Live-cell imaging
Live-cell imaging of ER, Golgi and the plasma membrane was conducted using the Axiovert135TV epifluorescence microscope (Zeiss) with 63× oil objective (Zeiss) and Olympus epifluorescence microscope with 40× oil objective and an additional 1.6× magnification. Twenty-four hours after transfection, cells were imaged by the CCD camera (QImaging) driven by Metamorph 7.5 imaging software (Molecular Devices) at 15 second time interval for at least 15 minutes. Mitochondria were imaged on the Leica SP5 inverted confocal microscope, with resonant scanner, HCX PL APO cS 40× objective lens, NA = 1.25 at 15 second time interval for at least 15 minutes. Mitochondria were also imaged on the spinning-disc Axiovert 200 confocal microscope (Zeiss). YFP and mCherry excitations were conducted with an argon laser (CVI-Melles Griot) with 40× objective (Zeiss) and an additional 1.6× magnification and NA = 1.30. 100 nM of rapamycin (Tecoland) was added after approximately 2 min and 30 seconds of imaging.
Electron microscopy
COS-7 cells were seeded in 35 mm dishes and transfected with Lyn-FRB and GFP-FKBP-N-BAR, Lyn-FRB and YFP-FKBP-NBAR, or Lyn-FRB and GFP-FKBP-ΔN-BAR using Amaxa Nucleofector (Lonza) or FuGENE HD (Roche) for transfection. Following 24–48 hours after transfection, rapamycin was added to a final concentration of 100 nM and incubated for 10 minutes at 37°C. Cells were then fixed with 4% paraformaldehyde and 0.1% glutaraldehyde for 15 minutes at room temperature. Next, cells were incubated in 3% paraformaldehyde, 1.5% glutaradehyde in 0.1 M sodium cacodylate, 5 mM magnesium chloride, 2.5% sucrose pH 7.4 at room temperature for 1 hour. Samples were then washed in 0.1 M sodium cacodylate, 2.5% sucrose pH 7.4, 3 times, 5 minutes each. Samples were stored at 4°C in 100 mM sodium cacodylate until further processing for electron microscopy.
Cells were harvested and pelleted. Subsequently, the cell pellets were trimmed into 1 mm3 pieces; cryoprotected by infiltration with 2.3 M sucrose/30% polyvinyl pyrrolidone (10,000 MW)/PBS (pH 7.4) for 2 hr; and mounted onto cryo-pins and rapidly frozen in liquid nitrogen. Ultrathin cryosections were cut on a Leica UCT ultramicrotome equipped with an FCS cryo-attachment and collected onto formvar/carbon-coated copper grids. The grids were then washed through several drops of 1X PBS containing 2.5% fetal calf serum (FCS) and 10 mM glycine (pH 7.4); then blocked in 10% FCS for 30 min; and incubated overnight in Chicken anti-GFP diluted 1:400 (Product # ab13970, Abcam, Cambridge, MA 02139). After washing, the grids were incubated for 2 hr in Donkey anti-chicken 12 nm Au (Jackson ImmunoResearch, West Grove, PA, USA 19390). The grids were washed through several drops of PBS followed by several drops of ddH2O; floated on a 1 ml drop of neutral UA, pH 7.4 for 10 min; quickly washed through five drops of ddH2O; and floated onto an aqueous solution containing 3.2% polyvinyl alcohol (10,000 MW), 0.2% methyl cellulose (400 centiposes) and 0.1% uranyl acetate. The grids were then embedded by removing excess solution using #50 hardened Whatman filter paper; and examined in a FEI (Hillsboro, OR, USA) Tecnai 12 Twin TEM operating at 100 kV. Images were collected using an Olympus Soft Imaging System (Lakeland, CO, USA) Megaview III digital camera.
Quantification of organelle morphological phenotypes
Morphological phenotypes were visually quantified using epifluorescence microscopy, as described below, in Metamorph 7.5 imaging software. Linear adjustments in brightness/contrast of epifluorescence images were adjusted using Metamorph and ImageJ 1.44 and 5× zoomed insets were obtained using ImageJ 1.44 “Zoom in images and stacks action tool” macro by Gilles Carpentier. Electron microscopy images analyzed using Olympus iTEM software and ImageJ. Linear adjustments in brightness/contrast of EM images were made using Adobe Photoshop.
The quantification of BAR-induced morphological phenotypes was conducted by counting the frequency of identified phenotypes occurring in the cell population after rapamycin treatment. All phenotypes were quantified with respect to the FRB-anchor images, relative to the organelle morphology prior to rapamycin treatment. All quantification was based on morphological changes occurring within 15 minutes to observe immediate effects of BAR recruitment. Filopodia (3.1 to 5.1 μm in length) and tubulations (2.1 to 33.6 μm in length, extending or retracting at a rate at least 0.1 μm/sec) at the plasma membrane were visually counted and the frequency of cells displaying these characteristics was recorded. Cells displaying rapid or unusual swinging mitochondrial movement greater than 0.2 μm/sec were included as BAR-induced phenotypes. BAR recruitment to the ER resulted in punctate structures along the tubular and sheet-like network, ranging in size from 1 to 60 μm2. Additional experiments recruiting the ER to the plasma membrane, with CFP-FKBP-Cb5 and Lyn-YFP-FRB constructs, respectively, were conducted to compare this fusion phenotype to BAR-induced phenotypes at the ER. At the cis- and medial-Golgi complex where YFP-FRB-Giantin or mCherry-FRB-Giantin localizes, protrusive or tubular structures ranging from 0.2 to 1.9 μm in width at a speed of less than 0.01 μm/sec were recorded as the phenotype. These protrusive structures differed from the naturally occurring dynamic tubules occurring at a rate greater than 0.1 μm/sec with a width less than 0.2 μm11.
Quantification of GFP-FKBP-N-BAR and GFP-FKBP-ΔN-BAR electron microscopy images was conducted by computing the number of anti-GFP immunogold particles along intracellular membranes. The number of gold particles within 100 nm of the plasma membrane surface was determined and compared to total labeling of the surface and internal membrane of the cell. Additionally, internal membrane tubules were identified and the number of gold particles within 100 nm of the tubules was calculated for the N-BAR and ΔN-BAR negative control. Since gold-labeling of N-BAR occurs on a greater length scale than N-BAR alone, analysis was conducted with respect to the length 100 nm. This encompasses the summation of lengths of the following components: FKBP, FRB, GFP, N-BAR or ΔN-BAR, chicken anti-GFP (IgY) and donkey anti-chicken 12 nm gold particle.
Statistical analysis
Statistical analysis of epifluorescence images was performed on a total of at least 30 cells from 3 independent trials. Mean +/− SD was recorded in Table 1. P values were calculated from unpaired, two-tailed Student's t-tests. Experimental data sets were compared to corresponding mutant BAR and FKBP data sets. Statistical analysis of EM images was performed by computing Mean +/− SD of immunogold labeling of intracellular membranes. P values were calculated from unpaired, two-tailed Student's t-tests of images clearly depicting plasma membrane (8 images analyzed for each N-BAR or ΔN-BAR conditions comparing plasma membrane surface labeling with total labeling of surface and internal membranes) or clearly showing membrane tubulation (14 images analyzed for N-BAR or ΔN-BAR conditions comparing immunogold labeling along internal membrane structures). P values were recorded in the Fig. 3 legend. P values less than 0.01 were considered statistically significant.
References
Shibata, Y. et al. Mechanisms shaping the membranes of cellular organelles. Ann Rev Cell Dev Biol. 25, 329–354 (2009).
McMahon, H. T. & Gallop, J. L. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature. 438, 590–596 (2005).
Peter, B. et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 303, 495–499 (2004).
Lee, E. et al. Amphiphysin 2 (Bin 1) and T-Tubule Biogenesis in Muscle. Science. 297, 1193–1196 (2002).
Hatzakis, N. S. et al. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nature Chemical Biology. 5, 835–841 (2009).
Frost, A. et al. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2, 191–196 (2009).
Frost, A. et al. Structural Basis of Membrane Invagination by F-BAR Domains. Cell. 132, 807–817 (2008).
Saarikangas, J. et al. Molecular mechanisms of molecular deformation by I-BAR domain proteins. Current biology. 19, 95–107 (2009).
DeRose, R. et al. Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 465, 409–417 (2013).
Komatsu, T. et al. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nature Methods. 7, 206–208 (2010).
Presley, J. F. et al. Golgi Membrane Dynamics. Molecular Biology of the Cell. 9, 1617–1626 (1998).
Bigay, J. et al. Curvature, Lipid Packing and Electrostatics of Membrane Organelles: Defining Cellular Territories in Determining Specificity. Developmental Cell. 23, 886–895 (2012).
Zimmerberg, J. & McLaughlin, S. Membrane Curvature: How BAR Domains Bend Bilayers. Current Biology. 14, R250–R52 (2004).
Lyman, E. et al. Water under the BAR. Biophysical Journal. 99, 1783–1790 (2010).
Mattila, P. K. & Lappalainen, P. Filopodia: molecular architecture and cellular functions. Nature Reviews Mol Cell Bio. 9, 446–454 (2008).
Lackner, L. L. & Nunnari, J. M. The molecular mechanism and cellular functions of mitochondrial division. Biochimica et Biophysica Acta (BBA) – Molecular Basis of Disease. 1792, 1138–1144 (2009).
Voeltz, G. et al. Structural organization of the endoplasmic reticulum. EMBO Reports. 3, 944–950 (2002).
Acknowledgements
We are grateful for the constructs encoding human Amphiphysin 1 N-BAR and human FBP17 F-BAR from Dr. Toshiki Itoh (Kobe University) and human MIM I-BAR from Dr. Pekka Lappalainen (University of Helsinki). We also extend our gratitude to Dr. T. Komatsu, R. DeRose and T. Sharma for constructive comments on the manuscript. This study was supported in part by the National Institute of Health (NIH) (GM092930 to T.I.) and by JST (10216 to T.I.).
Author information
Authors and Affiliations
Contributions
A.S. and T.U. performed cell biology experiments described in Figures 1 and 2, as well as in Table 1. J.M.M. conducted electron micrograph experiments shown in Figure 3. R.H. made some of the DNA constructs. A.S. and T.I. wrote the manuscript, which was reviewed by all authors.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Infomation
Supplementary Information
Supplementary Movie 1
Supplementary Information
Supplementary Movie 2
Supplementary Information
Supplementary Movie 3
Supplementary Information
Supplementary Movie 4
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images in this article are included in the article's Creative Commons license, unless indicated otherwise in the image credit; if the image is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the image. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Suarez, A., Ueno, T., Huebner, R. et al. Bin/Amphiphysin/Rvs (BAR) family members bend membranes in cells. Sci Rep 4, 4693 (2014). https://doi.org/10.1038/srep04693
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep04693
This article is cited by
-
Defunctionalizing intracellular organelles such as mitochondria and peroxisomes with engineered phospholipase A/acyltransferases
Nature Communications (2022)
-
Phospholipid flippase ATP11C is endocytosed and downregulated following Ca2+-mediated protein kinase C activation
Nature Communications (2017)
-
Membrane tubule formation by banana-shaped proteins with or without transient network structure
Scientific Reports (2016)
-
Membrane tension controls the assembly of curvature-generating proteins
Nature Communications (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.