Abstract
Emergence of drug-resistant strains of the pathogen Mycobacterium tuberculosis (Mtb) and the ineffectiveness of BCG in curtailing Mtb infection makes vaccine development for tuberculosis an important objective. Identifying immunogenic CD8+ T cell peptide epitopes is necessary for peptide-based vaccine strategies. We present a three-tiered strategy for identifying and validating immunogenic peptides: first, identify peptides that form stable complexes with class I MHC molecules; second, determine whether cytotoxic T lymphocytes (CTLs) raised against the whole protein antigen recognize and lyse target cells pulsed with peptides that passed step 1; third, determine whether peptides that passed step 2, when administered in vivo as a vaccine in HLA-A2 transgenic mice, elicit CTLs that lyse target cells expressing the whole protein antigen. Our innovative approach uses dendritic cells transfected with Mtb antigen-encoding mRNA to drive antigen expression. Using this strategy, we have identified five novel peptide epitopes from the Mtb proteins Apa, Mtb8.4 and Mtb19.
Similar content being viewed by others
Introduction
Based on the 2013 World Health Organization (WHO) report, Mycobacterium tuberculosis (Mtb) infection caused tuberculosis (TB) in 8.6 million people and 1.3 million deaths in 2012 and remains a major health concern in the developing world and among HIV-positive people. This situation is further exacerbated by the emergence of drug-resistant strains, multidrug-resistant (MDR), extensively drug-resistant (XDR) and totally drug-resistant strains1,2. Patients infected with these drug-resistant Mtb strains require much longer treatment courses and have lower success rates compared to susceptible strains3,4. No truly effective vaccine to prevent infectious TB exists5,6,7,8. Bacillus Calmette-Guérin (BCG) is a live attenuated vaccine used in some parts of the world, but due to its limited efficacy it is not routinely utilized in the United States9. Although there have been numerous vaccine studies to date, there is not a consensus as to the best antigens to include in a potential subunit vaccine against TB5,6,7,8,10.
A cellular CD8+ T cell immune response against Mtb requires that peptides derived from Mtb proteins be presented in the context of MHC class I molecules on Mtb-infected cells. These peptides are specifically recognized by CD8+ cytotoxic T lymphocytes (CTL) that lyse the infected cell. Current data in both humans and the murine model of TB infection indicate that CD8+ T lymphocyte responses are crucial to immunity against Mtb11,12,13,14,15,16,17,18,19. The antigen-specific CD8+ T cells lyse Mtb-infected monocyte-derived macrophages and alveolar macrophages after recognition of mycobacterial antigens presented in the context of MHC class I molecules. Thus, identification of M. tuberculosis peptides that bind to class I alleles is critical for understanding the immune response to TB infection and for the development of effective vaccines.
To generate an effective peptide-based vaccine it is critical to identify peptides that not only stimulate a peptide-specific T cell response, but also represent biologically derived peptides that are presented by pathogen-infected cells20,21. This would ensure that a peptide vaccine would stimulate a peptide-specific T cell response that will ultimately lyse the infected target cells. In a review22, Lauemoller et al. concluded that peptide binding to MHC is the single most selective event involved in antigen handling and the most important to predict. Since no peptide can become an epitope without first binding to MHC, a more detailed understanding of peptide binding to MHC molecules would facilitate the identification of potentially immunogenic T cell epitopes. Epitope mapping studies that have utilized peptide binding prediction algorithms have already identified HLA-A*0201 restricted epitopes in the Mycobacterium proteins ESAT-617,23, CFP10, CFP2124, 85B25, 85A26, 19 kDa lipoprotein, 16 kDa, 44 kDa, thymidylate synthase11, RNA poly B-subunit (RpoB)11, permease protein A-111, 38 kDa27, 28 kDa27 and other proteins28,29.
For the results reported herein, we employed a high throughput epitope discovery technology available through Beckman Coulter, Inc. (BCI) named iTopia™ that is optimized for use in identifying HLA class I associated epitopes. This technology does not rely on in silico algorithmic predictions, but instead provides for rapid experimental testing of all possible peptides and their bona fidein vitro binding characteristics to a broad group of the most common class I MHC alleles (A*0101, A*0201, A*2402, A*0301, A*1101, B*0702, B*0801 and B*1501). Our overall study objective toward a vaccine design for Mtb was to identify novel T cell epitopes capable of binding with high affinity and slow off-rates to HLA alleles and then to test the selected epitopes for immunogenicity. For this study, we examined three Mtb antigens (Apa, Mtb8.4 and Mtb19) all previously reported to be immunogenic30,31,32,33,34,35,36, but that have not been finely mapped for potential T cell epitopes.
Apa, a 50–55-kDa protein (Rv1860 gene) of M. tuberculosis33, has been shown to induce IFN-γ secretion from CD8+ T cells isolated from individuals with latent TB, as defined by a positive response to Purified Protein Derivative (PPD). In guinea pigs an Apa-specific vaccine consisting of DNA priming followed by a poxvirus recombinant booster injection resulted in protection against a virulent M. tuberculosis challenge32. Mtb8.4, a small protein present in the culture filtrate of M. tuberculosis, elicited Th1 responses in murine and human cells30,31. Notably, class I responses against this protein were demonstrated in PBMCs from PPD-positive humans30,31,36. Mtb8.4-specific T cells were detected in mice immunized with Mycobacterium bovis-BCG and in mice infected with M. tuberculosis30. More importantly, mice were protected from challenge with virulent M. tuberculosis and also demonstrated a strong induction of CD8+ cytotoxic T lymphocyte (CTL) responses after immunization with either plasmid DNA encoding Mtb 8.4 or Mtb 8.4 recombinant protein30. Finally, Mtb19 is a 19-kDa secreted lipoprotein that is another relevant target of cellular immunity to M. tuberculosis in the murine model of TB infection16,18. Although targeting Apa, Mtb8.4 and Mtb19 holds promise, to date very few studies have systematically addressed identification and functional validation of HLA-binding epitopes in Apa, Mtb8.434 and Mtb1935 proteins for any of the human alleles.
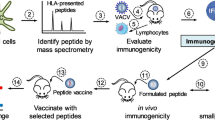
In this study we determined the immunogenicity of peptide epitopes that were selected based on optimal MHC class I binding properties. We then evaluated whether the cytotoxic T cells specific for the Mtb antigen were capable of recognizing and lysing cells that expressed the peptide-HLA complex. These CTL were generated in vitro using autologous dendritic cells (DCs) transfected with RNA encoding the Mtb antigen37,38,39,40. Mtb antigen-encoding RNA transfected DCs were used to induce Mtb antigen-specific CTL in vitro. In this study, we identified peptide epitopes by determining the ability of peptide-pulsed targets to be recognized and lysed by Mtb antigen-specific CTL. Peptides that were potential CTL epitopes, based on recognition and lysis of peptide-pulsed targets, were then used to stimulate antigen-specific T cells in vivo in HLA-A2 transgenic mice.
Results
Identifying peptide epitopes using iTopia™
We mapped and validated MHC class I-restricted T cell epitopes of Mtb antigens, using as our primary mapping tool iTopia™41,42,43, a Beckman Coulter, Inc. (BCI) technology. We analyzed overlapping 9-mer peptides (with 8 aa overlaps) that spanned the entire protein sequence of seven different M. tuberculosis antigens: CFP-10, ESAT6, Ag85A, Ag85B, Mtb19, Apa and Mtb8.4. We identified potential peptide epitopes among these 9-mers via a sequence of screening steps, such that only peptides that passed a given step were moved forward to the subsequent step. We performed an initial screening for MHC class I binding to one of eight alleles (A*0101, A*0201, A*2402, A*0301, A*1101, B*0702, B*0801 and B*1501) using the iTopia™ system. The binding measurements produced by iTopia™ along with the affinity (ED50) and off-rate (t1/2) measurements of peptide binding to the MHC class I molecules, were used to calculate the peptide's cScore (described in Methods and Supplementary Methods). The set of peptides with cScores above a 0.1 threshold were identified as the 9-mers with highest potential for being peptide epitopes and were moved into the functional validation phase of study. The cumulative result of peptide mapping for the 3 Mtb proteins of interest (Apa, Mtb8.4 and Mtb19) is provided in Table 1. The peptide mapping data for all the seven M. tuberculosis antigens: CFP-10, ESAT6, Ag85A, Ag85B, Apa, Mtb8.4 and Mtb19 is provided in Supplementary Table 1.
Generation of Mtb antigen-specific CTL responses using DCs transfected with RNA encoding Mtb antigen to stimulate autologous T cells in vitro
Although we identified a large number of potential epitopes that met the selection criteria (1396 9-mers for all seven TB proteins and 8 MHC alleles, see Supplementary Table 1), we focused on the functional validation of the epitopes in three of the lesser studied Mtb antigens, namely Apa, Mtb8.4 and Mtb19 (reducing the number of 9-mers to 570 for 8 MHC alleles, Table 1). Moreover, since HLA-A2 and HLA-B7 mice are available for functional validation, we further narrowed our analysis to HLA-A*0201- and HLA-B*0702-binding 9-mers from Mtb antigens, Apa, Mtb8.4 and Mtb19. Since our analysis was restricted to the top 20 peptides with cScore > 0.1, we analyzed a total of 75 peptides (57 HLA-A*0201- and 18 HLA-B*0702-binding peptides) using the validation strategy described in the results below.
To determine if peptides we identified are presented and recognized by antigen-specific CTL, our first task was to generate Mtb antigen-specific T cells in vitro that could then be used in subsequent experiments to functionally screen Mtb peptides. To this end, we have developed an in vitro system wherein we can routinely generate primary, antigen-specific cytotoxic T cells using DCs transfected with RNA encoding the full-length protein44,45 to stimulate autologous, naïve T cells in vitro37,38,39,40. We, therefore, generated in vitro transcribed (IVT) RNA encoding Apa, Mtb8.4 and Mtb19. Figure 1 demonstrates that DCs transfected with Mtb antigen RNA stimulate primary, antigen-specific CTL in vitro. The specificity of the lysis is demonstrated by the fact that Mtb antigen-specific effectors lyse only the corresponding Mtb antigen-expressing targets and not the control targets (DCs transfected with human actin RNA). Mtb antigen-specific CTL generated using this DC-RNA T cell stimulation platform were used to functionally screen peptides from Table 1.
Induction of Mtb antigen-specific CTL using Mtb antigen encoding RNA-transfected DCs to stimulate autologous T cells.
RNA-transfected mature DCs were co-cultured with autologous negatively isolated CD3+ T cells as described in Methods. Following in vitro stimulation, T cells were harvested and assessed for their ability to lyse targets in a standard 4-hour europium-release assay using autologous DCs transfected with RNA as targets. Standard deviation reflects variation between triplicate wells. This experiment was done 2 times with similar results. E:T is effector to target ratio.
Identification of endogenously processed and presented MHC class I HLA-A*0201-restricted and MHC class I HLA-B*0702-restricted peptides
The antigen-specific CTL generated using the protocol described in Figure 1 were used to determine immunodominance among the peptide epitopes that were identified using our screening algorithm. We determined whether the HLA-A*0201-restricted CTL epitopes predicted using the binding, affinity and off-rate algorithm were indeed endogenously processed peptides. Thus, peptide-pulsed target cells were tested for recognition and lysis by antigen-specific CTL22,46. The CTL assay also determines if the peptides with higher cScore are more efficiently recognized by Mtb antigen-specific T cells. CTL reactivity was determined by monitoring lysis of target cells, T2 cell line (TAP-deficient, HLA-A*0201+ cells) (Figure 2) or autologous DCs from HLA-A*0201+ donors (Figure 3) that were pulsed with peptides. Target cells pulsed with control peptides (depicted as C) were used as negative controls to determine the specificity of lysis. The peptides listed in Figures 2 and 3 are ranked in descending order from the highest to the lowest cScore binders. The peptide ID number refers to the nomenclature of the peptide and represents the starting amino acid position within the protein. This analysis was repeated using cells derived from 3 additional HLA-A*0201+ donors to determine reproducibility in the immunodominance of the top 20 cScore-ranked peptide epitopes. Each experiment is tabulated in Supplementary Table 2: 2a (Apa), 2b (Mtb8.4) and 2c (Mtb19). The sequence of peptides that were consistently recognized by antigen-specific CTL and their cScore ranking is provided in Table 2.
Identification of HLA-A*0201-restricted epitopes endogenously processed and presented by Mtb antigen encoding RNA-transfected DCs using T2 cells as targets.
Mtb antigen-specific T cells were generated as described in Methods. Europium-labeled T2 cells were incubated with the peptides (100 μM) for 1 h prior to addition of T cells followed by a standard 4 h europium release assay. Negative control (C) is an HLA-A*0201 peptide with a low cScore. Standard deviation reflects variation between triplicate wells. This experiment was done 3 times and data from each experiment is shown in Supplementary Table 2a–c. (2A). Mtb Apa, (2B). Mtb8.4, (2C). Mtb19.
Identification of HLA-A*0201-restricted epitopes endogenously processed and presented by Mtb antigen encoding RNA-transfected DCs using DCs as targets.
Mtb antigen-specific T cells were generated as described in Methods. Europium-labeled DCs were incubated with the peptides (100 μM) for 1 h prior to addition of T cells followed by a standard 4 h europium release assay. Negative control (C) is an HLA-A*0201 peptide with a low cScore. Standard deviation reflects variation between triplicate wells. This experiment was done 2 times and data from each experiment is shown in Supplementary Table 2a–c. (3A). Mtb Apa, (3B). Mtb8.4, (3C). Mtb19.
To functionally validate HLA-B*0702-binding peptide epitopes, HLA-B*0702-restricted T cells specific to Mtb antigens were generated in vitro using cells from healthy HLA-B*0702+ donors. These HLA-B*0702-restricted T cells were used in CTL assays with autologous DCs pulsed with peptide as targets (Figure 4a). To further validate the HLA-B*0702-binding peptide epitopes, we used an alternate approach. HLA-B7 transgenic mice were immunized subcutaneously three times with autologous DCs transfected with Mtb antigen-encoding RNA. Antigen-specific T cells were restimulated in vitro with RNA-transfected DCs and used to validate HLA-B*0702-restricted CTL reactivity using autologous DCs pulsed with peptides as targets (Figure 4b). As shown in Figure 4 and Table 2, both approaches revealed the presence of two dominant epitopes, one from Mtb antigen Apa and the second from Mtb antigen 19. The analysis was restricted to peptides with cScore > 0.1, therefore the analysis included 14 peptides from Apa, 3 peptides from Mtb8.4 and 1 peptide from Mtb19. This experiment was repeated 4 times using cells from 2 donors and cells from HLA-B7 mice and each experiment is represented in the Supplementary Table 2d.
Identification of HLA-B*0702-restricted epitopes for Mtb Apa, Mtb 8.4 and Mtb 19.
(A). Human CD3+ T cells were stimulated with autologous DCs transfected with either Mtb Apa RNA or Mtb 8.4 RNA or Mtb 19 RNA. Stimulations were as described in methods. Autologous human DCs pulsed with B*0702-restricted peptides were used as targets. Europium-labeled DCs were incubated with peptides (100 μM) for 1 h prior to addition of T cells followed by a standard 4 h europium-release assay. (B). HLA-B7 transgenic mice were immunized with murine DCs transfected with Mtb RNA 3 times at weekly interval. Splenocytes were harvested 8–9 days later and stimulated as described in methods. CTL assay was done after 5 days with murine DCs as targets. Europium-labeled DCs were incubated with the peptides (100 μM) for 1 h prior to addition of T cells followed by a standard 4 h europium release assay. Standard deviation reflects variation between triplicate wells. This experiment was done 4 times and data from each experiment is shown in Supplementary Table 2d. Peptides analyzed were based on cScore 0.1 or above (14 peptides for Mtb Apa, 3 peptides for Mtb 8.4 and 1 peptide for Mtb 19). Negative controls (C Apa, C 8.4 and C 19) are HLA-B*0702 peptides with a low cScore.
In vivo immunogenicity of peptide epitopes
HLA-A*0201-restricted peptides (2 peptides selected for each Mtb protein, Table 2) that were consistently recognized in CTL analysis in vitro were analyzed for immunogenicity by measuring their ability to induce a T cell response in vivo. HLA-A2 mice were immunized intradermally as indicated in the Methods with 100 μg of peptide, emulsified 1:1 in IFA, for a total of 3 immunizations in conjunction with anti-CD40 antibody. Non-adherent splenocytes were restimulated in vitro using peptide-pulsed DCs. CTL reactivity of these epitope-specific T cells was determined by measuring lysis of Mtb antigen-encoding RNA-transfected DCs as shown in Figure 5. Data in Figure 5 demonstrates that 5 out of the 6 HLA-A*0201-restricted peptides that were selected based on binding (Table 1) and functional screening (Figures 2 & 3), when administered in vivo in HLA-A2 transgenic mice as a vaccine, elicit cytotoxic T cells that lyse target cells expressing the Mtb antigen.
In vivo immunogenicity of Mtb peptides in HLA-A2 transgenic mice.
Mice were immunized intradermally 3 times at 1-week intervals with HLA-A*0201 peptides emulsified in IFA as described in detail in Methods. Peptides (2 representing each Mtb protein) were chosen based on in vitro assays shown in Figures 2 and 3 and Table 2. 10 days after the third immunization, spleens were harvested and non-adherent splenocytes were restimulated with autologous DCs pulsed with peptide and 50 U/ml IL-2. CTL assay was done 5 days later using the corresponding Mtb RNA-transfected DCs as targets. Actin RNA-transfected DCs were used as control targets. Standard deviation reflects variation between triplicate wells. E:T = 40:1.
Discussion
A number of methods have been proposed for identification of T cell epitopes based on the ability to predict or measure peptide binding to MHC class I molecules47,48,49. Most algorithmic approaches are limited to assessment of the theoretical potential for epitope binding in common alleles and do not measure actual binding or address the potential significant contribution of immunogenic epitopes in other HLA alleles. The methods that do measure binding are not amenable to high throughput and, therefore, it is difficult to screen large numbers of peptides for many different MHC types. The iTopia™ technology addresses these issues43. Using a modified iTopia™ algorithm to accurately assess binding, affinity and off-rate, we mapped and validated MHC class I-restricted T cell epitopes within selected target regions of lesser-studied Mtb antigens. Of note is the fact that the identified epitopes in Mtb have a proclivity for restriction by B*0702 and B*1501, as previously shown34, in addition to A*0201 (Table 1 and Supplementary Table 1). This higher restriction by B alleles may be suggestive of their potential immunogenicity in populations where HLA-B MHC alleles predominate.
To immunologically assess the function of these peptide epitopes, something that no algorithm is currently able to do, we generated Mtb antigen-specific T cells using DCs transfected with Mtb antigen-encoding RNA, technology that we originally established to stimulate tumor-specific immune responses in vivo37,38,39,40. Use of antigen-encoding mRNA offers an important and not fully appreciated, practical advantage over the use peptide or protein: ease of generation. Given a sequence, the corresponding mRNA can be generated in a simple process involving RT-PCR and transcription, with cloning of the cDNA intermediate into bacterial plasmid. Thus, the analysis of multiple antigenic targets is not hindered by the need to manufacture protein antigen or to identify class I and class II peptides corresponding to specific MHC alleles.
In this study, we identified functional peptide epitopes by determining the ability of peptide-pulsed targets to be recognized and lysed by Mtb antigen-specific CTL. Peptides that were potential CTL epitopes, based on recognition and lysis of peptide-pulsed targets, were then used to stimulate antigen-specific T cells in vivo in HLA transgenic mice. Our unique approach has identified Mtb peptide epitopes that are preferentially recognized by Mtb antigen-specific T cells. Notably, peptides with high binding scores were not necessarily the peptides recognized by Mtb-antigen-specific T cells, suggesting that detailed measurements of peptide-MHC binding (ED50 and t1/2) as well as functional analysis are both important in identifying immunogenic peptide epitopes. In follow-up studies, we will conduct ex vivo validation of HLA A*0201 epitopes of TB peptides identified in this study, using PBMCs from patients infected with Mtb followed by in vivo studies to determine if HLA-A2 mice vaccinated with these epitopes are protected against a virulent Mtb challenge. Although broad applicability of peptide vaccines for global diseases may be limited because of MHC restrictions and low immunogenicity, these limitations can be resolved by making multiple epitope vaccines, including CD4 T cell epitopes and with the use of adjuvants or chemical modifications to increase immunogenicity7,8. Furthermore, this study is also relevant in the selection of antigenic targets (immunogenic pathogen proteins) that can be incorporated into vaccine strategies (vector- or protein-based) designed to induce protective CD8+ T cell responses5,8,12.
It is also noteworthy that we did not see correlation between iTopia™ and the SYFPETHI and Bimas algorithms (peptide rankings compared in Supplementary Table 2), indicating that regardless of the algorithm used to identify MHC class I binding peptides, a functional validation should be performed to ultimately identify immunogenic T cell epitopes. The workflow for identification and validation of immunologically relevant peptides is shown in Supplementary Figure 1.
To summarize, the immunogenic peptide epitopes selected by binding, affinity and off-rate measurements followed by functional validation are listed below.
Mtb Apa: HLA-A*0201-binding peptides 25 (aa SASLVTVAV), 197 (aa YMPYPGTRI) and 263 (aa WLGTANNPV) and HLA-B*0702-binding peptide 286 (aa LVAPPPAPA) are potentially immunogenic epitopes. Positive lysis of T2 cell targets but not DC targets (refer to Supplementary Table 2) is presumably because T2 cells are artificially designed to exclusively present a high number of HLA-A*0201-binding epitopes thus increasing the density of these peptide epitopes on the cell surface. Moreover DCs express multiple HLA alleles and the individual alleles may differ in the levels of expression. We also noticed differences in peptide recognition between the different donors, although most donors were selected based on the expression of HLA-A*0201 and HLA-B*0702. In confirmatory in vivo immunogenicity assays, both peptides tested, peptides 25 and 263, induced immune responses in HLA-A2 transgenic mice (Figure 5).
Mtb 8.4: HLA-A*0201-binding peptides 4 (aa SLTALSAGV), 75 (aa APPPQRAAM), 82 (aa AMAAQLQAV) and 2 (aa RLSLTALSA) are potentially immunogenic epitopes. None of the 3 HLA-B*0702-restricted peptide epitopes that were selected using iTopia™ were immunogenic using our functional validation criteria. In confirmatory in vivo immunogenicity assays, both peptides tested, peptides 4 and 75, induced immune responses in HLA-A2 transgenic mice (Figure 5).
Mtb 19: HLA-A*0201-binding peptides 108 (aa VTLGYTSGT), 131 (aa KITGTATGV) and HLA-B*0702-binding peptide 146 (aa SPVNKSFEI) are potentially immunogenic epitopes. Although peptide 131 (aa KITGTATGV) was a strong candidate in functional assays using in vitro validation criteria, this peptide gave minimal CTL responses when used as an immunogen in HLA-A2 transgenic mice (Figure 5). Mtb 19 peptide 108 did induce a response in vivo in HLA-A2 transgenic mice (Figure 5).
Overall, we observed a weak correlation between the strength of peptide-MHC binding (as measured via iTopia™ as well as computed by prediction algorithms) and immunogenicity (as measured using functional lysis assays). This was also the case for two highly immunogenic peptides derived from melanoma tumor antigen MART (EAAGIGILTV) and influenza matrix protein M1 (GILGFVFTLT). These peptides elicit immunodominant HLA-A*0201 restricted T cell responses in vitro but do not have the highest predicted binding to HLA-A*0201 when compared to other peptides derived from the protein (data not shown).
While minimal binding between peptide and MHC is a requirement for immunogenicity, it alone is not sufficient or predictive of immunogenicity. Hence, peptide-MHC binding assessment serves as a first screening step to eliminate primarily true negative peptides, that is, peptides that most likely will fail to elicit an effective CTL response because they fail to be presented by MHC class I molecules. This pre-screening step is essential in the process of identification of peptide epitopes because experimental assays to assess immunogenicity are both costly and time-consuming; as such, it is unrealistic to perform such assays for large numbers of peptides. For these reasons, iTopia™ epitope discovery system (as possibly, in silico prediction algorithms) is a useful tool in the identification of peptide epitopes. In conclusion, in addition to identifying possible peptide epitopes for inclusion in peptide vaccine strategies, this immunological validation study may aid in the development of a more reliable algorithm for prediction of immunogenic epitopes that elicit potent MHC class I-restricted CD8+ T cell responses.
Methods
Ethics statement
Human
Primary human cells used in these experiments were isolated from leukapheresis products that were obtained from human subjects following informed written consent using protocols approved by the Duke University Institutional Review Board.
Murine
In conducting the research described in this paper, the investigators adhered to the “Guide for the Care and Use of Laboratory Animals” as proposed by the committee on care of Laboratory Animal Resources Commission on Life Sciences, National Research Council. The facilities at the Duke vivarium are fully accredited by the American Association for Accreditation of Laboratory Animal Care and all studies were conducted using protocols approved by the Duke University IACUC.
Peptide synthesis
All peptides (8 amino acids (aa), 9 aa and 10 aa) were synthesized at 80% or higher purity as assessed by HPLC and NMR profiling for each peptide (JPT Peptide Technologies). Peptides were dissolved in DMSO to generate 0.01 M stock solutions. The stock solutions were diluted in AIM-V to 0.001 M (10−3 M) and used in assays at 100 μM.
MHC class I binding, affinity and off-rate assays
Eight different Human Leukocyte Antigen (HLA) alleles that cover 85–90% of the world population were assessed (A*0101, A*0201, A*2402, A*0301, A*1101, B*0702, B*0801, B*1501). MHC molecules were produced using recombinant technology by Beckman Coulter Inc. Peptide-MHC class I binding, affinity and off-rate were measured using iTopia™ epitope discovery system as follows. Solid phase MHC class I molecules were biotinylated and bound to avidin coated 96-well microtiter plates. Class I devoid of peptide is unstable, so the bound class I molecule included a placeholder peptide to maintain its integrity. The complexes were denatured to release placeholder peptide and free β2 microglobulin and peptide were washed away. The test peptide, additional β2 microglobulin and antibody were added. The anti-HLA class I monoclonal antibody (mAb) only recognizes a properly folded class I molecule that contains peptide. To test affinity, peptides were incubated in decreasing concentrations (8 serial dilutions: 10−4, 10−8 M) for 18 hours. Results are expressed as the concentration of peptide required to achieve 50% binding or half the complexes to form (ED50). The entire plate was used to estimate common parameters of the affinity assay. To determine off-rate: peptide-MHC I complexes were allowed to form (18 hours) as in binding assay. Rate of dissociation from MHC was measured by loss of fluorescence over 8 different times (0, 0.5, 1.0, 1.5, 2.0, 4.0, 6.0, 8.0 hours) at 37°C. Results are expressed as the amount of time needed to achieve 50% dissociation (t1/2) of peptide from MHC complex. Positive controls were included on every plate. Each positive control value was within 3STDEV of the mean for all targets as tracked throughout the project with Levey-Jennings plots (data not shown). Plates were read on a plate reader with a fluorescent read-out (M2, Molecular Devices). This information was used to derive the binding score (or cScore) for each peptide (Table 1, Supplementary Table 1 and Supplementary Methods).
Cloning of genes
Dr. Richard Frothingham (Duke University) provided chromosomal DNA from Mtb H37Rv (ATCC 25681, type strain). PCR primers were designed based on the genomic sequence (GenBank AL123456) to amplify the Mtb genes for Apa (GenBank CCP44626; synonyms: Rv1860, antigen MPT-32, ModD), Mtb8.4 (GenBank NP_215690; synonym: Rv1174c) and Mtb19 (Genbank CCP46590; synonyms: Rv3763, LpqH, 19 kDa lipoprotein). Full-length Mtb genes were then sub-cloned into the RNA expression plasmid. The RNA expression plasmid is a derivative of pcDNA3.1(−) (Invitrogen) containing a T7 RNA polymerase site 5′ to the insertion site and a string of 64 adenosine residues 3′ to the insert. In vitro RNA transcription from this plasmid produces RNA with poly-A tail 64 bases in length. Successful sub-cloning of each gene was confirmed by DNA sequencing of each plasmid (data not shown).
In vitro transcription of RNA
The plasmids were digested with the restriction enzyme SpeI to linearize the DNA followed by in vitro transcription using the T7 mMessage Machine (Ambion). The in vitro transcribed (IVT) RNA was purified using a commercial kit (RNeasy, Qiagen), quantified by spectrophotometry and analyzed by MOPS formaldehyde gel electrophoresis to confirm the synthesis of full-length RNA (data not shown)44.
Generation of human dendritic cells (DCs)
Peripheral blood mononuclear cells (PBMCs) were thawed, washed in PBS and resuspended at 2 × 108 cells in 30 ml AIM-V media (Invitrogen) in T-150 tissue culture flasks45. Cells were incubated for 1 hour at 37°C, 5% CO2 in a humidified incubator. The non-adherent cells were removed by rocking the flask from side to side to dislodge them. The adherent cells were replenished with 30 ml AIM-V media with 800 U/ml human GM-CSF and 500 U/ml human IL-4 then incubated at 37°C. DCs were harvested on day 6, by collecting all non-adherent cells, followed by cold PBS wash. DCs that were still adherent were dissociated with cell dissociation buffer (Invitrogen), 37°C for 20 min. The DCs were washed, counted and maintained on ice until use.
Transfection of DCs with RNA encoding Mtb proteins
DCs were washed two times and gently resuspended in Opti-MEM at 2.5–3 × 107/ml. 100–200 μl were transferred to a 2-mm cuvette for electroporation using an Electro Square Porator ECM 830 from BTX® Harvard Apparatus, Inc (Holliston, MA). 3–4 μg RNA per 106 cells was added to the cuvette and mixed by pipetting up and down gently but thoroughly. The cells and RNA were pulsed at 300 Volts for 500 microseconds. The efficiency of RNA transfection was assessed using RNA encoding green fluorescent protein (GFP) followed by flow cytometric analysis (data not shown)44. Transfected DCs were immediately transferred to plates containing media at 106 cells/ml and incubated in media containing the maturation cytokine cocktail at 37°C, 5% CO2 for 8–15 hours. Maturation media is AIM-V containing GM-CSF (800 U/ml), IL-4 (500 U/ml), TNF-α (5 ng/ml), IL-1β (5 ng/ml), IL-6 (150 ng/ml) and PGE2 (1 μg/ml). All cytokines are obtained from Peprotech Inc. PGE2 is purchased from Sigma.
Stimulation of T cells with RNA-transfected DCs in vitro
Human DCs transfected with antigen (or control)-encoding RNA were used to stimulate T cell responses in vitro. PBMCs were thawed, washed, resuspended in PBS and treated with DNase I (Sigma) at 200 U/ml for 20 min at 37°C. The EasySep T cell Enrichment Kit (StemCell Technologies, Vancouver, Canada) was used to negatively isolate CD3+ T cells from DNase I-treated PBMCs. The isolated CD3+ T cells were stimulated with RNA-transfected, matured DCs at a responder to stimulator ratio (R:S) of 10:1 in the presence of 25 ng/ml IL-7 at 37°C. Stimulations were done in RPMI 1640 with 10% FCS, 2 mM L-glutamine, 20 mM HEPES, 1 mM Na-pyruvate, 0.1 mM MEM non-essential amino acids, MEM amino acids (50× solution), 100 IU/ml penicillin, 100 μg/ml streptomycin and 5 × 10−5 M β-mercaptoethanol (CTL stimulation medium). The responder T cell concentration was 2–2.5 × 106 cells/ml. IL-2 was added on day 3 at 50–100 U/ml. On day 7, the T cells were restimulated with RNA-electroporated DC at a R:S ratio of 10:1 in complete RPMI-10% FCS in the presence of 50–100 U/ml IL-2, with the T cells at 1–2 × 106 cells/ml in CTL stimulation medium. After 5–7 days the T cells were harvested, counted and used as effector cells in a europium-based CTL assay50.
Target cells for CTL assays
T2 cells pulsed with peptides or autologous DCs pulsed with peptides or transfected with RNA were used as targets. T2 (ATCC® CRL-1992™) is an HLA-A*0201+ T-B lymphoblastoid hybrid cell line deficient of transporter associated with antigen processing (TAP) proteins and therefore can be successfully loaded with the candidate peptide epitopes51. DCs were transfected with RNA and incubated overnight in GM-CSF+IL-4 AIM-V media prior to europium labeling for use as target cells. 104 europium-labeled target cells in 100 μl of CTL medium (without penicillin-streptomycin) were incubated with 100 μM of peptide per well in 96-well V-bottom plates for 1 hour at 37°C. Effector cells were added to these peptide-pulsed, europium-labeled target cells for CTL assay at the indicated effector to target (E:T) ratio.
In vitro cytotoxicity assay
5–10 × 106 target cells were labeled with europium for 20 min at 4°C using a standard published procedure50. 104 europium-labeled targets and serial dilutions of effector cells at varying E:T ratio or at fixed E:T ratio as indicated in the figure legends were incubated in 200 μl of CTL medium (CTL stimulation medium with no penicillin-streptomycin) in 96-well V-bottom plates. The plates were centrifuged at 500 × g for 3 min and incubated at 37°C for 4 hours. 50 μl of the supernatant was harvested and added to 150 μl of enhancement solution (Perkin-Elmer) in 96-well flat-bottom plates and europium release was measured by time resolved fluorescence using the VICTOR3 Multilabel Counter (Perkin-Elmer).
Specific cytotoxic activity was determined using the formula: % specific release = [(experimental release - spontaneous release)/(total release - spontaneous release)] × 100. Spontaneous release of the target cells was less than 25% of total release by detergent in all assays for the results of the assay to be valid. Spontaneous release of the target cells was determined by incubating target cells in medium without T cells. Non-specific lysis was determined by measuring the lysis of target cells loaded with control peptides (C). All assays were done in triplicates and repeated 2–3 times to confirm data.
Mice
4–6 week old C57Bl/6 HLA-A2 transgenic mice (Stock #004191, Jackson Labs) and C57Bl/6 HLA-B7 transgenic mice (STOCK Tg(HLA-B*0702/H2-Kb)B7.xx β2mtm1UncTg(β2M)55Hpl, Taconic) were used. The HLA-A2 (HLA-A*0201) transgenic mice express α1, α2 domain of HLA-A*0201, α3 domain of H-2Dd and human β2M (β2 microglobulin). The HLA-B7 (HLA-B*0702) transgenic mice express α1, α2 of HLA-B*0702, α3 domain of H-2Kb and human β2M.
Generation of murine bone marrow precursor-derived DCs
Marrow from tibias and femurs of mice were harvested followed by treatment with ammonium chloride Tris buffer for 3 min at 37°C to deplete the red blood cells38,52. The precursors were plated in RPMI-5% FCS with GM-CSF (15 ng/ml) and IL-4 (10 ng/ml) and supplemented with 20 mM HEPES, 2 mM L-glutamine, 1 mM sodium-pyruvate, 100 IU/ml penicillin, 100 mg/ml streptomycin, 0.1 mM non-essential amino acids and 5 × 10−5 M β-mercaptoethanol in 6-well plates. Cells were maintained at 106/ml, 3 ml/well and incubated at 37°C and 5% CO2. After 3 days the floating cells (mostly granulocytes) were removed and the adherent cells replenished with fresh GM-CSF and IL-4 containing medium. 4 days later the non-adherent DCs were harvested, washed, pulsed with peptide (100 μM) or electroporated with RNA as described above and matured for 5 hours at 37°C in GM-CSF+IL4 RPMI-5% FCS medium supplemented with LPS (E. coli 026:B6, 100 ng/ml, Sigma).
CTL induction in vivo
Naïve mice were immunized intradermally, 3 times at 12-day intervals. Each immunization consisted of an emulsion containing 100 μg of peptide, 1:1 with incomplete Freund's adjuvant in a total volume of 100 μl per injection. Mice were also injected with 50 μg of anti-CD40 (eBioscience) given subcutaneously on day 0 and day 1 of each immunization. Alternately, mice were immunized subcutaneously, at the base of each ear, 3 times at 7–10 day intervals with peptide-pulsed or RNA-transfected DCs (2–3 × 105 DCs per mouse in 100 μl PBS, 50 μl per site).
Splenocytes were harvested 8–10 days after the third immunization and depleted of red blood cells with ammonium chloride Tris buffer. Splenocytes were adhered for 1 hour in tissue culture plates to deplete adherent cells. 107 non-adherent splenocytes were cultured with 2 × 105 stimulator cells (DCs pulsed with corresponding peptides used for immunization or Mtb RNA-transfectd DCs) in 5 ml of RPMI with 10% FCS, 20 mM HEPES, 2 mM L-glutamine, 1 mM sodium pyruvate, 100 IU/ml penicillin, 100 μg/ml streptomycin, 0.1 mM non-essential amino acids and 5 × 10−5 M β-mercaptoethanol (complete RPMI-10% FCS) per well and 50 U/ml of IL-2 in a 6-well tissue culture plate at 37°C and 5% CO2. Effectors were harvested 5 days later on Histopaque 1083 gradient (Sigma) for harvesting viable lymphocytes prior to use in a CTL assay (described above). DCs transfected with Mtb antigen-encoding RNA or pulsed with Mtb peptides were used as targets.
References
Dye, C., Glaziou, P., Floyd, K. & Raviglione, M. Prospects for Tuberculosis Elimination. Annu Rev Public Health 34, 271–286, 10.1146/annurev-publhealth-031912-114431 (2012).
Gandhi, N. R. et al. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet 375, 1830–1843, 10.1016/S0140-6736(10)60410-2 (2010).
Muller, B., Borrell, S., Rose, G. & Gagneux, S. The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet 29, 160–169, 10.1016/j.tig.2012.11.005 (2012).
Smith, T., Wolff, K. A. & Nguyen, L. Molecular Biology of Drug Resistance in Mycobacterium tuberculosis. Curr Top Microbiol Immunol 374, 53–80, 10.1007/82_2012_279 (2012).
Behar, S. M., Woodworth, J. S. & Wu, Y. Next generation: tuberculosis vaccines that elicit protective CD8+ T cells. Expert Rev Vaccines 6, 441–456, 10.1586/14760584.6.3.441 (2007).
Kaufmann, S. H. Future vaccination strategies against tuberculosis: thinking outside the box. Immunity 33, 567–577, 10.1016/j.immuni.2010.09.015 (2010).
Gowthaman, U., Rai, P. K., Khan, N., Jackson, D. C. & Agrewala, J. N. Lipidated promiscuous peptides vaccine for tuberculosis-endemic regions. Trends Mol Med 18, 607–614, 10.1016/j.molmed.2012.07.008 (2012).
Ottenhoff, T. H. & Kaufmann, S. H. Vaccines against tuberculosis: where are we and where do we need to go? PLoS Pathog 8, e1002607, 10.1371/journal.ppat.1002607 (2012).
Andersen, P. & Doherty, T. M. The success and failure of BCG - implications for a novel tuberculosis vaccine. Nat Rev Microbiol 3, 656–662, 10.1038/nrmicro1211 (2005).
Prabowo, S. A. et al. Targeting multidrug-resistant tuberculosis (MDR-TB) by therapeutic vaccines. Med Microbiol Immunol 202, 95–104, 10.1007/s00430-012-0278-6 (2012).
Cho, S. et al. Antimicrobial activity of MHC class I-restricted CD8+ T cells in human tuberculosis. Proc Natl Acad Sci U S A 97, 12210–12215 (2000).
Grotzke, J. E. & Lewinsohn, D. M. Role of CD8+ T lymphocytes in control of Mycobacterium tuberculosis infection. Microbes Infect 7, 776–788, 10.1016/j.micinf.2005.03.001 (2005).
Lancioni, C. et al. CD8+ T cells provide an immunologic signature of tuberculosis in young children. Am J Respir Crit Care Med 185, 206–212, 10.1164/rccm.201107-1355OC (2012).
Nagata, T. & Koide, Y. Induction of Specific CD8 T Cells against Intracellular Bacteria by CD8 T-Cell-Oriented Immunization Approaches. J Biomed Biotechnol 2010, 764542, 10.1155/2010/764542 (2010).
Caccamo, N. et al. Analysis of Mycobacterium tuberculosis-specific CD8 T-cells in patients with active tuberculosis and in individuals with latent infection. PLoS One 4, e5528; 10.1371/journal.pone.0005528 (2009).
Klein, M. R. & McAdam, K. P. CD8+ T lymphocytes against mycobacterium tuberculosis. Arch Immunol Ther Exp (Warsz) 47, 313–320 (1999).
Lalvani, A. et al. Human cytolytic and interferon gamma-secreting CD8+ T lymphocytes specific for Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 95, 270–275 (1998).
Vordermeier, H. M., Zhu, X. & Harris, D. P. Induction of CD8+ CTL recognizing mycobacterial peptides. Scand J Immunol 45, 521–526 (1997).
Axelsson-Robertson, R. et al. Extensive major histocompatibility complex class I binding promiscuity for Mycobacterium tuberculosis TB10.4 peptides and immune dominance of human leucocyte antigen (HLA)-B*0702 and HLA-B*0801 alleles in TB10.4 CD8 T-cell responses. Immunology 129, 496–505, 10.1111/j.1365-2567.2009.03201.x (2010).
Flynn, J. L. & Chan, J. Immunology of tuberculosis. Annu Rev Immunol 19, 93–129, 10.1146/annurev.immunol.19.1.93 (2001).
Axelsson-Robertson, R., Magalhaes, I., Parida, S. K., Zumla, A. & Maeurer, M. The immunological footprint of Mycobacterium tuberculosis T-cell epitope recognition. J Infect Dis 205 Suppl 2, S301–315, 10.1093/infdis/jis198 (2012).
Lauemoller, S. L. et al. Identifying cytotoxic T cell epitopes from genomic and proteomic information: “The human MHC project”. Rev Immunogenet 2, 477–491 (2000).
Aagaard, C. S., Hoang, T. T., Vingsbo-Lundberg, C., Dietrich, J. & Andersen, P. Quality and vaccine efficacy of CD4+ T cell responses directed to dominant and subdominant epitopes in ESAT-6 from Mycobacterium tuberculosis. J Immunol 183, 2659–2668, 10.4049/jimmunol.0900947 (2009).
Lv, H. et al. Identification of a novel cytotoxic T lymphocyte epitope from CFP21, a secreted protein of Mycobacterium tuberculosis. Immunol Lett 133, 94–98, 10.1016/j.imlet.2010.07.007 (2010).
Geluk, A. et al. Identification of major epitopes of Mycobacterium tuberculosis AG85B that are recognized by HLA-A*0201-restricted CD8+ T cells in HLA-transgenic mice and humans. J Immunol 165, 6463–6471 (2000).
Smith, S. M. et al. Human CD8+ CTL specific for the mycobacterial major secreted antigen 85A. J Immunol 165, 7088–7095 (2000).
Shams, H., Barnes, P. F., Weis, S. E., Klucar, P. & Wizel, B. Human CD8+ T cells recognize epitopes of the 28-kDa hemolysin and the 38-kDa antigen of Mycobacterium tuberculosis. J Leukoc Biol 74, 1008–1014 (2003).
Tang, S. T. et al. Genome-based in silico identification of new Mycobacterium tuberculosis antigens activating polyfunctional CD8+ T cells in human tuberculosis. J Immunol 186, 1068–1080, 10.4049/jimmunol.1002212 (2011).
Chen, F. et al. In vitro and in vivo identification of a novel cytotoxic T lymphocyte epitope from Rv3425 of Mycobacterium tuberculosis. Microbiol Immunol 56, 548–553, 10.1111/j.1348-0421.2012.00470.x (2012).
Coler, R. N. et al. Vaccination with the T cell antigen Mtb 8.4 protects against challenge with Mycobacterium tuberculosis. J Immunol 166, 6227–6235 (2001).
Coler, R. N. et al. Molecular cloning and immunologic reactivity of a novel low molecular mass antigen of Mycobacterium tuberculosis. J Immunol 161, 2356–2364 (1998).
Kumar, P., Amara, R. R., Challu, V. K., Chadda, V. K. & Satchidanandam, V. The Apa protein of Mycobacterium tuberculosis stimulates gamma interferon-secreting CD4+ and CD8+ T cells from purified protein derivative-positive individuals and affords protection in a guinea pig model. Infect Immun 71, 1929–1937 (2003).
Laqueyrerie, A. et al. Cloning, sequencing and expression of the apa gene coding for the Mycobacterium tuberculosis 45/47-kilodalton secreted antigen complex. Infect Immun 63, 4003–4010 (1995).
Lewinsohn, D. A. et al. Immunodominant tuberculosis CD8 antigens preferentially restricted by HLA-B. PLoS Pathog 3, 1240–1249, 10.1371/journal.ppat.0030127 (2007).
Mohagheghpour, N. et al. CTL response to Mycobacterium tuberculosis: identification of an immunogenic epitope in the 19-kDa lipoprotein. J Immunol 161, 2400–2406 (1998).
Skeiky, Y. A. et al. Cloning, expression and immunological evaluation of two putative secreted serine protease antigens of Mycobacterium tuberculosis. Infect Immun 67, 3998–4007 (1999).
Nair, S. K. et al. Induction of primary carcinoembryonic antigen (CEA)-specific cytotoxic T lymphocytes in vitro using human dendritic cells transfected with RNA. Nature Biotechnology 16, 364–369 (1998).
Nair, S. K. et al. Induction of cytotoxic T cell responses and tumor immunity against unrelated tumors using telomerase reverse transcriptase RNA transfected dendritic cells.[comment]. Nature Medicine 6, 1011–1017 (2000).
Nair, S. K. et al. Induction of tumor-specific cytotoxic T lymphocytes in cancer patients by autologous tumor RNA-transfected dendritic cells. Annals of Surgery 235, 540–549 (2002).
Thornburg, C., Boczkowski, D., Gilboa, E. & Nair, S. K. Induction of cytotoxic T lymphocytes with dendritic cells transfected with human papillomavirus E6 and E7 RNA: implications for cervical cancer immunotherapy. Journal of Immunotherapy 23, 412–418 (2000).
Bachinsky, M. M. et al. Mapping and binding analysis of peptides derived from the tumor-associated antigen survivin for eight HLA alleles. Cancer Immun 5, 6 (2005).
Shingler, W. H., Chikoti, P., Kingsman, S. M. & Harrop, R. Identification and functional validation of MHC class I epitopes in the tumor-associated antigen 5T4. Int Immunol 20, 1057–1066, 10.1093/intimm/dxn063 (2008).
Wulf, M., Hoehn, P. & Trinder, P. Identification of human MHC class I binding peptides using the iTOPIA- epitope discovery system. Methods Mol Biol 524, 361–367, 10.1007/978-1-59745-450-6_26 (2009).
Lee, J., Boczkowski, D. & Nair, S. Programming human dendritic cells with mRNA. Methods Mol Biol 969, 111–125, 10.1007/978-1-62703-260-5_8 (2013).
Nair, S., Archer, G. E. & Tedder, T. F. Isolation and generation of human dendritic cells. Curr Protoc Immunol Chapter 7, Unit7.32, 10.1002/0471142735.im0732s99 (2012).
Deng, Y., Yewdell, J., Eisenlohr, L. & Bennink, J. MHC affinity, peptide liberation, T cell repertoire and immunodominance all contribute to the paucity of MHC class I-restricted peptides recognized by antiviral CTL. J Immunol 158, 1507–1515 (1997).
Larsen, M. V. et al. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency and proteasomal cleavage predictions. Eur J Immunol 35, 2295–2303, 10.1002/eji.200425811 (2005).
Rammensee, H., Bachmann, J., Emmerich, N. P., Bachor, O. A. & Stevanovic, S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 (1999).
Saethang, T. et al. EpicCapo: epitope prediction using combined information of amino acid pairwise contact potentials and HLA-peptide contact site information. BMC Bioinformatics 13, 313; 10.1186/1471-2105-13-313 (2012).
Blomberg, K., Granberg, C., Hemmila, I. & Lovgren, T. Europium-labelled target cells in an assay of natural killer cell activity. II. A novel non-radioactive method based on time-resolved fluorescence. significance and specificity of the method. J Immunol Methods 92, 117–123 (1986).
Salter, R. D. & Cresswell, P. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO J 5, 943–949 (1986).
Inaba, K. et al. Identification of proliferating dendritic cell precursors in mouse blood. J Exp Med 175, 1157–1167 (1992).
Acknowledgements
This work was entirely supported by the US NIH/NIAID award to K.W. (N01-AI-40082). The authors would like to thank Dr. Richard Frothingham for providing Mtb chromosomal DNA, Thomas Denny for providing healthy donor leukaphereses and David Snyder for help with murine studies.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: S.N., G.T. and K.W. Performed the experiments: S.N., D.B., G.T., A.S., K.P. and Y.C. Analyzed the data: S.N., G.T., K.W., D.B., A.S., K.P. and Y.C. Contributed reagents/materials/analysis tools: S.N., G.T., K.W., A.S., C.C., J.D., T.K. and S.P. Wrote the manuscript: S.N., K.W., A.S., D.B., S.P. and G.T. All authors reviewed the manuscript and approved the final version of the manuscript.
Ethics declarations
Competing interests
S.N. and D.B. are co-inventors on a patent that describes the use of dendritic cells transfected with tumor antigen encoding RNA that has been licensed by Argos Therapeutics (Durham, NC) through Duke University. S.N. and D.B. have no financial interests in Argos Therapeutics and are not compensated by Argos Therapeutics.
Electronic supplementary material
Supplementary Information
Supplementary Data
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images in this article are included in the article's Creative Commons license, unless indicated otherwise in the image credit; if the image is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the image. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Nair, S., Tomaras, G., Sales, A. et al. High-throughput identification and dendritic cell-based functional validation of MHC class I-restricted Mycobacterium tuberculosis epitopes. Sci Rep 4, 4632 (2014). https://doi.org/10.1038/srep04632
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep04632
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.