Abstract
Tomato and its processed products are one of the most widely consumed fruits. Its domestication, however, has resulted in the loss of some 95% of the genetic and chemical diversity of wild relatives. In order to elucidate this diversity, exploit its potential for plant breeding, as well as understand its biological significance, analytical approaches have been developed, alongside the production of genetic crosses of wild relatives with commercial varieties. In this article, we describe a multi-platform metabolomic analysis, using NMR, mass spectrometry and HPLC, of introgression lines of Solanum pennellii with a domesticated line in order to analyse and quantify alleles (QTL) responsible for metabolic traits. We have identified QTL for health-related antioxidant carotenoids and tocopherols, as well as molecular signatures for some 2000 compounds. Correlation analyses have revealed intricate interactions in isoprenoid formation in the plastid that can be extrapolated to other crop plants.
Similar content being viewed by others
Introduction
The cultivated tomato, Solanum lycopersicum, is one of the most widely consumed fruits. Over 150 million metric tons are produced annually, contributing to a $35 billion industry1. Many of the essential and beneficial nutrients in the human diet, such as antioxidants, vitamins and minerals, are derived from tomato fruit and its products2. In addition to being an economically important crop, it is also a model species, impacting on several areas of plant biology, such as fruit physiology and development3, quantitative genetics and plant breeding4,5. Over the last decade an impressive array of genetic resources for such studies has been developed, including populations of genetically defined inbreds, generated through crossing S. lycopersicum with wild relatives6, mutant collections7 and TILLING (targeting induced local lesions in genomes) platforms, generating a diverse range of mutants8. Procedures for the evaluation of protein composition9, enzyme activities10 and metabolites11 have also been developed. These multiple levels of analysis can now be exploited, following the sequencing of the tomato genome12 (http://solgenomics.net/), to enable a step-change in our ability to utilise the metabolic diversity and natural genetic variation of wild relatives for improving crop quality through metabolomics-assisted breeding13,14,15. It has been estimated that less than 5% of the genetic variation of wild relatives is present in domesticated species, due to selection of preferred genotypes in existing germplasm16, a process known as domestication syndrome17.
Metabolite profiling approaches have been used with several introgression (IL) populations, including that of the S. pennellii x S. lycopersicum, to evaluate chemical composition, identify quantitative trait loci (QTL) and facilitate their resolution18,19,20. In the present article, a complementary set of metabolomic approaches has been implemented to greatly extend existing metabolite data resources. Extracts of ripe fruit from the S. pennellii IL population have been analysed by NMR, positive (+ve) and negative (−ve) direct infusion mass spectrometry (Di-MS), high performance liquid chromatography with photodiode array detection (LC-PDA/MS) and gas chromatography-MS (GC-MS) and compared to the domesticated cultivar, M82. Identification of metabolite alleles in ILs, as well as computational integration of the datasets has been carried out and metabolic networks constructed for the tomato fruit metabolome and the individual pathway components. These data represent a valuable and timely resource to fully capitalise on the sequencing of tomato genomes. In addition, the applicability of tomato as a model for other crop plants means that the analytical approach developed in this study and data generated provide a generic resource.
Results
Introgression lines show significantly different metabolite profiles to M82
Principal component analysis (PCA) for the raw intensities of all variables yielded separation between the two seasonal crops for the ILs and M82 (Supplementary Figure S1). No defined classes of metabolites within a crop were responsible for clustering; instead a variety of intermediary metabolites were identified. Re-analysis of the dataset following normalization to the M82 control metabolites, however, indicated a high degree of co-localization of the genotypes to the same region of the scatter plot (Supplementary Figure S2). PCA of metabolites from each of the 76 ILs, using score and loadings plots, showed separation from the M82 parent (Supplementary Figure S3, A–L), indicating that in each IL the chemical composition was significantly altered by the presence of the introgressed genomic regions on each chromosome from the S. pennellii wild relative. In all cases, the variation could not be attributed to one metabolite.
To analyze individual metabolite perturbations throughout the metabolome, the putative metabolites, ascertained from the PlantCyc database, in combination with those confirmed through targeted metabolite profiling, were tabulated to show fold changes in molecular features relative to the M82 (Supplementary Dataset S1, ftp://ftp.solgenomics.net/projects/uk_metabolomics/Solanum_pennellii/) and categorized into over 600 pathways (Supplementary Dataset S2, ftp://ftp.solgenomics.net/projects/uk_metabolomics/Solanum_pennellii/). Representation in Cytoscape (http://www.cytoscape.org/) revealed metabolite changes exclusive to an IL, or those common to several ILs (Supplementary Figure S4, A–C), whilst chromosome maps, illustrating compounds that increase or decrease two fold relative to M82, are shown in Supplementary Figure S5, A–L. An overriding feature of the data is that no IL contains a perturbation in a single pathway or metabolite. The number of pathways affected within an IL can vary, as can the number of metabolites affected per pathway. For example, IL 11-4-1 has putative metabolite changes in only 389 out of 662 PlantCyc pathways (59%), the fewest shown, whereas all other ILs show changes in over 75% of the pathways. The pathways affected are often diverse, even within the same IL. Across the whole population the down-regulation of metabolites is slightly more predominant. Of the 20,309 statistically significant variable changes (p < 0.05), 10,377 were negative and 9,932 positive, but an 85% decrease in this number was found after false discovery rate (FDR) calculations (q < 0.05). A decrease or increase in metabolites is not exclusive to any one genotype, but across the population specific ILs have predominantly up or down regulation of metabolites. For example, of 321 altered variables (p < 0.05, q < 0.05) in IL 3-2, 291 are negative, whereas IL 7-4 has 364 positive changes out of 499 true altered variables (p < 0.05, q < 0.05). In total, 45 ILs have net metabolite increases, with 29 ILs showing net decreases of metabolites. Changes in metabolite levels that are common to multiple ILs can be observed within the data, with overlapping introgressed regions yielding discrete clusters of metabolites. These changes are not always restricted to overlapping regions, but can be found between ILs with no DNA regions in common.
Introgressions on chromosomes 3, 6, 8 and 12 are associated with changes to carotenoids and tocopherols
ILs 3-2, 6-3 and 12-2 have distinct fruit colour phenotypes, due to altered carotenoid pigment composition (Figures 1 to 3). The underlying candidate genes for these QTL are phytoene synthase (Psy-1), the chromoplast specific lycopene β-cyclase (B-CYC) and lycopene ε-cyclase (ε-Lcy), located on chromosomes 3, 6 and 12, respectively7. The loadings plots for each of these ILs, however, compared to the M82, indicate significant variation in other metabolites, not just those found in the carotenoid pathway. For example, the metabolites in IL 3-2 contributing to the overall variance included sucrose, in IL 6-3 glutamate and in IL12-2 adenosine monophosphate (Figures 1D to 3D). Metabolite profiling of ILs 8-2-1 and 8-2 revealed an increase in α-tocopherol and decrease in γ-tocopherol, with the overall total tocopherol levels elevated 2-fold (Figure 4C). To validate the identity of α and γ tocopherol and their amounts relative to M82, targeted LC-MS was performed. Co-chromatography of these compounds on several HPLC/UPLC systems and identical UV/Vis, MS and MS/MS spectra to that of the authentic standards enabled unambiguous identification of these tocopherols (Figure 4B). The scatter plots revealed both metabolites to be common to the two Chr8 ILs, whilst the physical map showed the levels of these two metabolites to be increased and decreased, respectively, in both 8-2-1 and 8-2 (Supplementary Figure S5H).
Metabolite changes associated with the altered colour phenotype of IL3-2.
(A) Carotenoid profiles, obtained by HPLC-UV/Vis analysis, recorded at 450 nm; (i), M82; (ii) IL3-2. The chromatographic peaks and UV/Vis spectra are 1-lutein, 2-β-carotene and 3-lycopene. (B) Quantitative changes associated with pathway components. The position of the proposed pathway block (the fruit- specific phytoene synthase-1) is indicated. (C) Physical map of chromosome 3, showing the position of IL3-2 and Psy-1. (D) PCA of the metabolomic dataset for IL3-2; (i), scatter diagram of the score values, highlighting the separation based on variance in chemical composition between IL3-2 (brown dots) and the M82 control (red dots). The % contribution of each component to the variance is shown (ii), loadings plot of variables with the identity of some of the variables annotated. Only variables contributing to a significant difference (p-value of < 0.05) are shown.
Metabolite changes associated with the altered colour phenotype of IL6-3.
(A) Carotenoid profiles, obtained by HPLC-UV/Vis analysis, recorded at 450 nm; (i), M82; (ii), IL6-3. The chromatographic peaks and UV/Vis spectra are 1-lutein, 2-β-carotene and 3-lycopene. (B) Quantitative changes associated with pathway components. The position of the proposed step in the pathway up-regulated (the fruit specific lycopene β-cyclase, CYC-B) is indicated. (C) Physical map of chromosome 6, showing the position of IL6-3 and CYC-B. (D) PCA of the metabolomic dataset for Il6-3; (i) scatter diagram of the score values, highlighting the separation based on variance in chemical composition between IL6-3 (brown dots) and the M82 control (red dots). The % contribution of each component to the variance is shown (ii), loadings plot of variables with the identity of some of the variables annotated. Only variables contributing to a significant difference (p-value of < 0.05) are shown.
Metabolite changes associated with the altered colour phenotype of IL12-2.
(A) Carotenoid profiles, obtained by HPLC-UV/Vis analysis, recorded at 450 nm; (i), M82; (ii), IL12-2. The chromatographic peaks and UV/Vis spectra are 1-lutein, 2-β-carotene and 3-lycopene. (B) Quantitative changes associated with pathway components. The position of the proposed up-regulation in the pathway (lycopene ε-cyclase, Lyc-E) is indicated. (C) Physical map of chromosome 12, showing the position of Il12-2 and Lyc-E. (D) PCA of the metabolomic dataset for IL12-2; (i), scatter diagram of the score values, highlighting the separation based on variance in chemical composition between IL12-2 (brown dots) and the M82 control (red dots).
The identification of candidate genes from the metabolomic dataset.
(A) Changes in molecular features (metabolites) associated with Chr8 ILs. Each green dot is an IL, red dot a molecular feature (putative metabolite). The black connecting lines show an increase, the blue a decrease. The cluster of features circled represents significant metabolite changes common to both 8-2 and 8-2-1, of which a reduction in γ-tocopherol and increase in α-tocopherol was observed. (B) LC-MS profile validating the identity of tocopherols. (C) Quantitative changes (± s.e.) in tocopherols (γ and α) found in IL8-2 and IL8-2-1, relative to their M82 comparator. (D) Co-localization of the candidate gene with the change in tocopherol metabolites in IL8-2 and 8-2-1, as well as the position of the candidate gene product within the tocopherol biosynthetic pathway.
Correlation networks for plastid isoprenoid formation reveal linkage of core, intermediary and tertiary metabolism
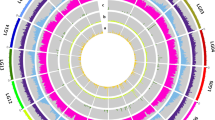
Correlation networks for metabolism occurring in ripening fruit have been built from the metabolomic databases. These networks have been constructed for individual genotypes and collectively across the population, using biochemical pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/). The strength of the networks ranged from 0.2 to 0.8, with an average of 0.45 (Figure 5). Positive and negative correlations occurred, with positive attributes predominating. The over-riding feature of the network is the grouping of primary metabolism in the centre of the network, with secondary metabolic components on the periphery, with just a few metabolites creating links to the core. Isoprenoid biosynthesis, which is responsible for the formation of vitamin E (tocopherols) and carotenoids, was used to demonstrate the utility of the dataset. The sectors of metabolism displaying strong associations to tocopherols and carotenoids are visualised in Figure 5.
A correlation network constructed from metabolites involved in chloroplast isoprenoid biosynthesis.
Pearson coefficients (r 0.6 to 0.8 with a p-value significance < 0.05) derived from the metabolite levels associated with all 76 ILs of the S. pennellii collection. Metabolites associated with the Calvin cycle are shown as blue dots, the chloroplast located 2-C-methyl-D-erythritol-4-phosphate (MEP) pathway purple dots, chlorophyll degradation green dots, phytyl synthesis khaki coloured, tocopherol red coloured, prenyl lipid biosynthesis yellow and carotenoid formation orange. Green connecting lines represent positive correlations and red connecting lines negative. Putative hubs derived from the number of connections are circled and the putative metabolites annotated. Collectively, the networks' strengths varied from 0.4 to 0.8 with the average being 0.45. Abbreviations: GGPP, geranylgeranyl diphosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; MEP, methylerythritol pathway.
Discussion
The effect on metabolite profiles of the cultivation of the same tomato population in two seasons is evident from the PCA, shown in Supplementary Figure S1. However, when the metabolite changes to the control (M82) line are used to normalize the data, then close clustering of the profiles occurs (Supplementary Figure S2). Thus, environmental/cultivation conditions, even in a glasshouse, cause changes in the chemical composition of the IL population. These environmental effects were minimized by normalizing the data, so that each variable per plant is relative to the mean of the variables from all control plants. Collectively, the statistical and multivariate analyses of the normalized data indicate that the dataset is robust and accurate and the effects of genetic determinants predominates over environmental influences, allowing metabolite perturbations caused solely by the introgression of the S. pennellii genome into M82 to be analysed for genetic determinants. A similar approach, using field-grown crops and focusing on hydrophilic metabolites, has been reported11, whilst a later investigation verified the heritability of QTL and the validity of this approach20. The changes in the metabolite composition of the ILs (Figures 1,2,3; Supplementary Figures S3–5; Supplementary datasets S1 and 2) may be due to a number of factors. These include alleles in a single gene (biosynthetic or regulatory), in multiple unrelated alleles, the result of changes in a metabolite or group of metabolites affecting other pathways, or processes via transcriptional or post transcriptional mechanisms, including variation in the kinetic properties of enzymes catalysing step(s) in metabolic pathways. In previous studies on genetically modified tomato fruit, we have found that post-transcriptional controls are important for changes to carotenoid levels, rather than gene expression21. The ability of the introgressed regions to modify whole pathways and processes suggests an important role for transcriptional regulators in the control of metabolism. The breadth and diversity of the changes would also suggest that regulation by small molecules must also not be overlooked. Although there is no correlation between the size of introgression and the number of metabolite changes occurring within it, those with the greatest number of changes are presumably due to regulators being present in the introgressed regions that have multiple roles. A recent report has shown that the regulatory gene ethylene response factor 6 (SIERF6) influences both carotenogenesis and additional ripening phenotypes22. So called master regulators are well documented in plant secondary metabolism23.
The large metabolomic datasets created in this study enable the identification of metabolites associated with important agronomic and consumer traits. Such comprehensive phenotyping is recognised for its diagnostic strength in genomics-assisted selection for crop improvement24,25. 2820 mQTL can be attributed to health-related traits, with 1,474 increased and 1,346 decreased levels, compared to M82. When used in combination with the genome sequence, the full potential of the present dataset will provide a direct route to candidate genes to test their commercial potential for improving the nutritional quality of crops. For example, ILs 3-2, 6-3 and 12-2 have changes to carotenoids, whilst ILs 8-2 and 8-2-1 exhibit increases in α-tocopherol (Figures 1,2,3,4). It is likely that the changes in product/precursor ratio of tocopherols relate to levels of γ-methyl tocopherol transferase, which is responsible for the conversion of γ-tocopherol to α-tocopherol and located at the end-point of the biosynthetic pathway (Figure 4D). The γ-methyl tocopherol transferase gene (SGN-U584511) is located within the overlapping region of 8-2-1 and 8-2 (Fig. 4D). Allelic variation in the S. pennellii γ-methyl transferase has been identified26. These data demonstrate the potential of the dataset, in combination with the genome sequence and other genetic resources, to rapidly assign traits to candidate genes, with the recent advent of Marker2 sequence facilitating identification27. Such a route to candidate genes underlying important QTL also helps validate and build confidence in the large-scale metabolomic approach.
Since the provision of essential nutrients, particularly antioxidants, in the human diet is a key attribute of tomato products, networks associated with health promoting phytochemicals were prepared and interrogated using the metabolite datasets (Supplementary Datasets 1 and 2). Several important, generic features, reflecting metabolism in general, can be deduced: (i), precursors common to multiple pathways act as important nodes in the network; (ii), the influence of cofactors (reductants) in metabolic pathways is important and appears to have been undervalued previously (iii), metabolism does not necessarily partition into primary and secondary types; (iv), the strong link between the utilisation of photosynthate and renewable formation and (v), the independence of the plastid-localized isoprenoid formation and specific classes of isoprenoid metabolism. For example, no strong connections were apparent between plastidial 2-C-methyl-D-erythritol-4-phosphate (MEP)-derived isoprenoids in tomato and those non-plastidic compounds formed via mevalonate. In ripe tomato fruit, therefore, the existence of isoprenoid precursor exchange to and from the plastid is unlikely, which is consistent with the previous experimental evidence of the independent modulation of the two pathways28. Whether associations exist between earlier intermediates in core metabolism and other organelles awaits further experimentation, as discussed recently29.
The biosynthesis of isoprenoids, such as tocopherols and carotenoids and the degradation of chlorophyll, also group independently (Figure 5). However, key intermediates that are common to these pathways display a high level of connectivity, suggesting they are key regulatory hubs. If the network data are interpreted from a predictive viewpoint for the design of metabolic engineering strategies, then geranylgeranyl diphosphate synthase (GGPP synthase), which is responsible for the biosynthesis of common C20 precursors used in carotenoid and tocopherol formation and isopentenyl diphosphate isomerase/dimethylallyl diphosphate isomerase (IPP/DMAPP isomerase), catalysing the synthesis of the universal C5 building blocks for all isoprenoids, would be important targets. In bacteria GGPP synthase(s) and IPP/DMAPP isomerase(s) have been amplified with notable increases in end-products30,31. Under stress conditions, IPP/DMAPP isomerase has shown positive effects on carotenoid biosynthesis32. These findings are supported by flux coefficients for these enzymes in ripening fruit28. In contrast, phytoene synthase has a high flux coefficient and numerous transgenic experiments have demonstrated the influence this biochemical step can have over the carotenoid pathway33. Interestingly, the correlation network places phytoene and its products at the edge of the network, with very few connections (Figure 5). Several other metabolites associated with biochemical steps amenable to metabolic engineering predominate in the same area, with few connections, such as β-carotene and γ-tocopherol, both of which have been used as precursors for enhancing high levels of valuable products in plant hosts34,35. Although the correlation networks suggest several common pathway precursors as metabolic hubs, this cannot be readily validated experimentally, because of the effect of regulatory processes. For example, GGPP synthase and IPP isomerase are members of multigene families36, where redundancy could occur in order to safeguard the essential nature of these biosynthetic steps through compensatory isoenzymes. To a degree, this highly tuned regulation can be observed from the way that carotenoid content can be elevated readily in plant tissues where their formation is not endogenous, compared to tissues with optimised synthesis and sequestration has evolved37.
From network analysis, the individual isoprenoid pathways are connected via sequential prenyl lipid and methylerythritol phosphate (MEP) predominating clusters to the Calvin-Benson cycle at the core (Figure 5). Traditionally, the terms primary and secondary metabolism are used to describe different types of cellular metabolism. The metabolic networks constructed in the present study, however, do not fit into this classical format. For isoprenoid biosynthesis in tomato fruit, the Calvin-Benson cycle is positioned at the core and associated via intermediary modules, represented by the MEP and prenyl lipid pathways, to a tertiary level, which includes tocopherol and carotenoid biosynthesis. Thus, three distinct levels: core, intermediary and tertiary occur. The Calvin-Benson cycle is at the core of the network in ripening fruit, yet this process is typically associated with photosynthetic tissues. Although tomato fruit are photosynthetically active during fruit development38, it is not a key aspect of their metabolism. Recently, the role of photosynthesis in fruit development has been questioned, with the belief that photosynthate is derived from vegetative material39. The integrative characterisation of transgenic tomato plants possessing fruit specific down regulation of DETIOLATED140 is another example where core metabolism, particularly carbon fixation, appears to be the progenitor of down-stream secondary, or in this case, tertiary pathways in tomato fruit. The metabolite of the Calvin cycle displaying the greatest connection was D- glyceraldehyde-3-phosphate, which is common to both the Calvin cycle and the MEP pathway. Collectively, these findings suggest that the plasticity of Calvin cycle components in tomato fruit warrant further investigation and the utilization of photosynthate carbon close to its source, without further resource allocation, could be a valuable strategy to explore as part of a combinatory engineering approach. Other metabolic links between isoprenoid formation and photosynthetic carbon flow have been revealed through metabolomic analysis of hemiterpenoid glycosides under nutrient deprivation41. Finally, cofactors/reductants appear to have an important influence over multiple pathways, presumably by direct action on biosynthetic enzymes, or perhaps via the modulation of redox state, as suggested for cross talk between cytosolic and plastidial isoprenoid formation42, as well as the dual role of plastidial terminal oxidase in carotenoid formation and chlororespiration43.
In summary, the data presented in this investigation represent one of the most comprehensive metabolomic studies performed in planta and have increased our understanding of the structure and dynamics of the isoprenoid pathway network, a necessity for biotechnological applications44. In combination with the recently published potato and tomato genome sequences, a valuable resource has been created to associate traits to metabolites and metabolites to candidate genes and regions of multiple QTL. We envisage that the resource will be used to extract leads for functional genomics and that the data will impact directly on plant breeding strategies, help elucidate underlying molecular mechanisms associated with traits and provide exploitable datasets for systems biology approaches.
Methods
Cultivation of tomato crops
The S. lycopersicum (M82) x S. pennellii IL population, obtained from the Tomato Genetics Resource Center (http://tgrc.ucdavis.edu/pennellii_ils.aspx), was grown in glasshouse conditions (16 h day length, day temp 20°C, night temp 18°C) over two seasons. Plants were grown in 7.5 litre pots of Levington M2 compost and irrigation supplemented with Vitax 214. The population was planted in four blocks, each containing 1 plant of the 76 S. pennellii ILs, with three M82 controls.
Experimental design, data acquisition, integration and statistical analyses
From four plants of each genotype, six ripe fruit (9 days post breaker, dpb) were harvested, pooled, freeze-dried and homogenized into a fine powder. Triplicate technical replicates for polar (water/methanol 4:1)) and non-polar (chloroform) extracts were prepared and subjected to NMR and direct infusion (Di)-MS (in both +ve and −ve mode). These approaches were complemented by targeted procedures using GC-MS, LC-MS and HPLC-PDA techniques for known phytochemicals28,33,40. Quality control samples were concurrently analysed. Following acquisition and normalization of the data, computational integration of the datasets was performed. A data matrix that combined the analytical outputs and facilitated statistical analysis of metabolites/molecular features for each IL, compared to its M82 control, was the objective of the data processing (Supplementary Dataset S1). To achieve this goal, the data generated were treated in a variety of ways. Firstly, unique chemical shifts were used to identify metabolites within the NMR fingerprints45. This resulted in 500 variables from the NMR spectra. For direct and LC-MS analysis, the m/z signatures were assigned nominal masses and ion correlations (including ion adducts). This treatment resulted in 852 features in the +ve mode and 948 in −ve mode. Variables generated in +ve (POS) and −ve (NEG) ionisation modes were annotated incrementally from 50 to 999 (e.g. POS_50). These values represented binned intensities of 1 Da, whereby POS_108, for example, represents the median of intensities between m/z 107.5 and 108.5. The NMR data were treated in a similar manner with 900 variables consisting of 0.01 δ bins from 0.505 to 9.495 δ, annotated for example as NMR_0.505, where NMR_3.455 would equal the sum of intensities between 3.450 δ and 3.460 δ. The amounts of compounds determined by targeted analysis were also entered into the data matrix. The molecular weights were used to designate putative formulae, which in turn were used to blast against the Plant and MetaCyc databases, generating putative metabolite identifications, which led to reaction and pathway assignments. Confirmatory targeted GC-MS and LC-PDA gave rise to a further 30 to 60 metabolites. In this case, retention times and spectral properties compared to authentic standards were used to provide unambiguous identifications. Multivariate analysis was initially performed to assess the overall variance in the population and identify crude changes in chemical composition. The putative annotations are based on the premise that if the compound were present in tomato and were to form an ion under the ionisation conditions used, then it would be found in the designated MS bin. Correlation analyses were performed using Pearson coefficients, between putative bin identities and targeted compounds from NMR and HPLC, which showed a high level of correlation (see Figure 5; p < 0.05) for sucrose and hexose (NMR v MS), rutin, naringenin and chlorogenic acid (HPLC v MS), glutamine, glutamate, asparagine and aspartate (NMR v MS and GC/MS v MS).
The data were reduced using PCA and in order to account for environmental effects, they were also normalised to their M82 controls. This was achieved by first calculating the mean M82 value for each variable in crop 1 and then crop 2. Then, the ratio for each biological replicate (plant) to mean M82 ratio was calculated. Plants derived from crop 1 (including the M82 biological replicates) were normalised to the mean of the M82 comparator for crop 1 and likewise with crop 2. The mean fold changes from M82 for each of the variables found in every IL were then calculated and both ANOVA and Student's t-tests performed to identify variables that were statistically significantly different (p < 0.05) from M82. False detection rates (FDR) for putative mQTL were calculated using the method of Storey and Tibshirani46. The data were shown to be normally distributed using the Kolmogorov-Smirnov (K-S) test for normal distribution and also visualized using several complementary approaches. Heat maps were produced in MatLab v. (http://www.mathworks.co.uk/index.html) and used to show the fold-change for each variable in every IL compared to the M82 comparator. An in-house Excel macro was used to plot putative biochemical pathway changes for each IL, compared to M82. The putative ions for each compound in every pathway of the PlantCyc database were listed with their mean fold-change from M2 for all ILs. A graph was then plotted for every pathway (Supplementary Dataset S2). Pearson correlation coefficients were calculated between the putative compound ions from the mean fold-change from M82 across all ILs within each pathway. An in-house Excel macro was used to compile files compatible with Cytoscape software (http://www.cytoscape.org/). Correlations that were statistically significant (p < 0.05, two-tailed test) were plotted in Cytoscape to show both positive and negative interactions. The networks for adjoining biochemical pathways were then merged to provide an overview of the total network in tomato fruit.
References
Food and Agriculture Organization of the United Nations. Tomato production. http://faostat.fao.org/ (2013). Date of access 15/10/13.
Spencer, J. P. E. et al. The genomic variation of the antioxidant potential of different toamot varieties. Free Radic. Res. 39, 1005–1016 (2005).
Giovannoni, J. Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 725–749 (2001).
Zamir, D. Improving plant breeding with exotic genetic libraries. Nat. Rev. Genet. 2, 983–989 (2001).
Lippman, Z. B., Semel, Y. & Zamir, D. An integrated view of quantitative trait variation using tomato interspecific introgression lines. Curr. Opin. Genet. Dev. 17, 545–552 (2007).
Eshed, Y. & Zamir, D. Introgressions from Lycopersicon pennellii can improve the soluble solids yield of tomato hybrids. Theoret. Appl. Genet. 88, 891–897 (1994).
Liu, Y. S. et al. There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotech. J. 1, 195–207 (2003).
Minoia, S. et al. A new mutant genetic resource for tomato crop improvements by TILLING technology. BMC Res. Notes 3, 69 (2010).
Faurobert, M. et al. Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Physiol. 143, 1327–1346 (2007).
Steinhauser, M. C. et al. Enzyme activity profiles during fruit development in tomato cultivars and Solanum pennellii. Plant Physiol. 153, 80–98 (2010).
Schauer, N. et al. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24, 447–454 (2006).
The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).
Fernie, A. R., Tadmor, Y. & Zamir, D. Natural genetic variation for improving crop quality. Curr. Opin. Pant Biol. 9, 196–202 (2006).
Fernie, A. R. & Schauer, N. Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet. 25, 39–48 (2009).
Bai, Y. & Lindhout, P. Domestication and breeding of tomatoes: what have we gained and what can we gain in the future? Ann. Bot. 100, 1085–1094 (2007).
Miller, J. C. & Tanksley, S. D. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor. Appl. Genet. 80, 437–448 (1990).
Frary, A. & Doganlar, S. Comparative genetics of crop plant domestication and evolution. Turk. J. Agric. For. 27, 56–69 (2003).
Tieman, D. et al. Tomato aromatic amino acids decarboxylases participate in synthesis of the flavour volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. USA 103, 8287–8292 (2006).
Fridmann, E., Carrari, F., Lin, Y. S., Fernie, A. R. & Zamir, D. Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305, 1786–1789 (2004).
Schauer, N. et al. Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20, 509–523 (2008).
Fraser, P. D. et al. Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids and intermediary metabolism. The Plant Cell 19, 3194–3211 (2007).
Lee, J. M. et al. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. The Plant Journal 70, 191–204 (2012).
Cannes do Nascimento, N. & Fett-Neto, A. G. Plant secondary metabolism and challenges in modifying its operation: an overview. Methods in Mol. Biol. 643, 1–13 (2010).
Fernie, A. R., Tadmor, Y. & Zamir, D. Natural genetic variation for improving crop quality. Curr. Opin. Plant Biol. 9, 196–202 (2006).
Fernie, A. R. & Schauer, N. Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet. 25, 39–48 (2009).
Almeida, J. et al. Genetic dissection of vitamin E biosynthesis in tomato. J. Exp. Bot. 62, 3781–3798 (2011).
Chibon, P.-Y., Schoof, H., Visser, R. G. F. & Finkers, R. Marker2sequence, mine your QTL regions for candidate genes. Bioinformatics 28, 1921–1922 (2012).
Enfissi, E. M. A. et al. Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotechnol. J. 3, 17–27 (2005).
Sweetlove, L. J. & Fernie, A. R. The spatial organization of metabolism within the plant cell. Annu. Rev. Plant Biol. 64, 723–746.
Ruther, A., Misawa, N., Böger, P. & Sandmann, G. Production of zeaxanthin in Escherichia coli transformed with different carotenogenic plasmids. Appl. Microbiol. Biotechnol. 48, 162–167 (1997).
Kajiwara, S., Fraser, P. D., Kondo, K. & Misawa, N. Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem. J. 324, 421–426 (1997).
Nakamura, A., Shimada, H., Masuda, T., Ohta, H. & Takamiya, K. Two distinct isopentenyl diphosphate isomerases in cytosol and plastid are differentially induced by environmental stresses in tobacco. FEBS Lett. 506, 61–64 (2001).
Fraser, P. D. et al. Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit specific manner. Proc. Natl. Acad. Sci. USA 99, 1092–1097 (2002).
Fujisawa, M. et al. Pathway engineering of Brassica napus seeds using multiple key-enzyme genes involved in ketocarotenoid formation. J. Exp. Bot. 60, 1319–1332 (2009).
Shintani, D. & DellaPenna, D. Elevating the vitamin E content of plants through metabolic engineering. Science 282, 2098–2100 (1998).
Bouvier, F., Rahier, A. & Camara, B. Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 44, 357–429 (2005).
Fraser, P. D. & Bramley, P. M. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 43, 228–265 (2004).
Buker, M., Schunemann, D. & Borchert, S. Enzymic properties and capacities of developing tomato (Lycopersicon esculentum L.) fruit plastids. J. Exp. Bot. 49, 681–691 (1998).
Lytovchenko, A. et al. Tomato fruit photosynthesis is seemingly unimportant in primary metabolism and ripening but plays a considerable role in seed development. Plant Physiol. 157, 1650–1663 (2011).
Enfissi, E. M. A. et al. Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. The Plant Cell 22, 1190–1215 (2010).
Ward, J. L., Baker, J. M., Llewellyn, A. M., Hawkins, N. D. & Beale, M. H. Metabolomic analysis of Arabidopsis reveals hemiterpenoid glycosides as products of a nitrate ion-regulated, carbon overflow. Proc. Natl. Acad. Sci. USA 108, 10762–10767 (2011).
Laule, O. et al. Crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100, 6866–6871 (2003).
Shahbazi, M., Gilbert, M., Laboure, A.-M. & Kuntz, M. Dual role of the plastid terminal oxidase in tomato. Plant Physiol. 145, 691–702 (2007).
Vranova, E., Conran, D. & Gruissem, W. Structure and dynamics of the isoprenoid pathway network. Mol. Plant 5, 318–333 (2012).
Ward, J. L., Baker, J. M. & Beale, M. H. Recent applications of NMR spectroscopy in plant metabolomics. FEBS J. 274, 1126–1131 (2007).
Storey, J. D. & Tibshirani, R. Statistical significance for genome-wide experiments. Proc. Natl. Acad. Sci. 100, 9440–9445 (2003).
Acknowledgements
We are grateful for financial support from the Biotechnology and Biological Sciences Research Council (BBSRC) Plant and Microbial Metabolomic Initiative (grant # BB/D00716X/1) and the ERA-NET Plant Genomics TomQML programme of the European Commission to T.W., G.B.S., C.G., P.M.B. and P.D.F. Prof. Dani Zamir (Hebrew University of Jerusalem) is thanked for original IL seed material and Ms Aimee Llewellyn for technical support in tissue and sample logistics. We also thank Dr Lukas Mueller, Boyce Thompson Institute for Plant Research, New York, for placing supplementary data onto the Sol Genomics (http://solgenomics.net/) ftp internet site.
Author information
Authors and Affiliations
Contributions
P.D.F. and G.B.S. designed the study; D.I.C., J.L.W. and M.H.B. carried out the NMR and DiMS; T.W. and C.K.G. carried out the HPLC; G.B.S cultivated the plants and L.P.-F., G.B.S., T.W., M.H.B., P.M.B. and P.D.F. analysed data and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Perez-Fons, L., Wells, T., Corol, D. et al. A genome-wide metabolomic resource for tomato fruit from Solanum pennellii. Sci Rep 4, 3859 (2014). https://doi.org/10.1038/srep03859
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03859
This article is cited by
-
Genotyping-by-sequencing and multilocation evaluation of two interspecific backcross populations identify QTLs for yield-related traits in pigeonpea
Theoretical and Applied Genetics (2020)
-
Identification of metabolite traits from the current metabolomic approaches
Theoretical and Experimental Plant Physiology (2019)
-
Development and use of chromosome segment substitution lines as a genetic resource for crop improvement
Theoretical and Applied Genetics (2019)
-
Bacterial wilt and canker of tomato: fundamentals of a complex biological system
Euphytica (2018)
-
Crop metabolomics: from diagnostics to assisted breeding
Metabolomics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.