Abstract
Skeletal muscle (SM) regeneration after injury is impaired by excessive inflammation. Particularly, the inflammatory cytokine tumour necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK) is a potent inducer of skeletal muscle wasting and fibrosis. In this study we investigated the role of Nrf2, a major regulator of oxidative stress defence, in SM ischemia/reperfusion (I/R) injury and TWEAK induced atrophy. We explored the time-dependent expression of TWEAK after I/R in SM of Nrf2-wildtype (WT) and knockout (KO) mice. Nrf2-KO mice expressed significant higher levels of TWEAK as compared to WT mice. Consequently, Nrf2-KO mice present an insufficient regeneration as compared to Nrf2-WT mice. Moreover, TWEAK stimulation activates Nrf2 in the mouse myoblast cell line C2C12. This Nrf2 activation inhibits TWEAK induced atrophy in C2C12 differentiated myotubes. In summary, we show that Nrf2 protects SM from TWEAK-induced cell death in vitro and that Nrf2-deficient mice therefore have poorer muscle regeneration.
Similar content being viewed by others
Introduction
Muscle wasting occurs with chronic diseases like cancer, diabetes or obstructive pulmonary disease. Several surgical procedures, especially in reconstructive surgery, likewise cause severe muscle trauma, which hampers the outcomes of such operations. Yet skeletal muscle functionality is important for mobility and quality of life. Published reports have shown how cytokines induce skeletal muscle wasting, but in spite of this, anti-inflammatory drug therapies were not sufficiently able to avert muscle wasting1. The tumour necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK) is currently receiving attention from several groups who have described its very potent wasting effect on skeletal muscle cells both in-vivo and in-vitro2,3. TWEAK expression has been shown to increase after muscle trauma and TWEAK-deficient mice likewise show better muscle regeneration after trauma4.
The transcription factor Nrf2 is a master regulator of oxidative stress that plays a central role in the induction of antioxidative and phase II enzymes5,6. Nrf2 is constitutively expressed and degraded by its inhibitor, Keap1, which allows a fast response to any occurrence of stress. By oxidative stress or by direct Nrf2-phosphorylation, Keap1 inhibition terminates and Nrf2 translocates to the nucleus, where it binds to a specific consensus sequence known as antioxidant response element (ARE). This element is located in the promoter region of hundreds of genes and these genes can be induced via Nrf2. Several studies have indicated that Nrf2-ARE signalling might also play a role in the attenuation of inflammatory processes such as those of autoimmune diseases or atherosclerosis7.
Recently, Ichihara et al. demonstrated that after hindlimb ischemia in mice, Nrf2-ablation enhances inflammation and increases expression of cytokines such as TNF-alpha and IL-68. Several other studies have focussed on correlations between increased cytokine expression in Nrf2-knockout mice9,10,11. As of yet no study has elucidated how Nrf2 and TWEAK might interact.
Here, we report that Nrf2-deficient mice express higher amounts of TWEAK and that Nrf2-knockout (KO) retards skeletal muscle regeneration after injury.
Results
Expression levels of TWEAK are increased after hindlimb ischemia in Nrf2-KO mice
Ischemia reperfusion injury (I/R) is a set-up during which tissue is cut off from perfusion for a certain amount of time and then subjected to a period of reperfusion. This tissue setting allows observation of regeneration in various tissues, e.g. nerves, blood vessels or myofibres. I/R is often used to cause skeletal muscle injury in mice.
To investigate TWEAK expression levels at different time intervals after ischemia, we measured mRNA levels (Fig. 1A), protein expression via ELISA (Fig. 1B) and we performed immunostaining of TWEAK (Fig. 1C–J). First we examined TWEAK expression in healthy Nrf2-WT and KO mice, neither of which showed a significant difference in TWEAK expression between the two groups (51.16 ± 12.78 vs. 89.99 ± 17.58 pg/ml) (Fig. 1A, B). 6 h after ischemia, TWEAK levels remained unchanged in injured Nrf2-WT mice (37,6 ± 27.12 pg/ml), whereas the injured Nrf2-KO mice already showed increased levels of TWEAK expression (108.2 ± 23,29 pg/ml). 96 h after ischemia both genotypes showed elevated TWEAK levels, with Nrf2-KO mice showing significantly higher levels of TWEAK as compared to Nrf2-WT mice (180.2 ± 61.36 vs. 73.63 ± 11.36 pg/ml). Two weeks after ischemia, TWEAK levels had returned to the baseline standard in both groups (41.96 ± 1.55 vs. 56.24 ± 26.48 pg/ml) again.
Nrf2-KO mice express higher levels of TWEAK after I/R injury.
(A) Expression of TWEAK-mRNA was measured by RT-PCR of hindlimb muscle homogenates after four hours of I/R. (B) Levels of TWEAK protein were measured by ELISA. (C–J) The cellular localization of TWEAK was detected using immunofluorence staining with TWEAK-antibodies performed on sections of healthy (C, D) and Nrf2-WT or KO mice, both at 6 h (E, F), 96 h (G, H) or 14 d (I, J) after I/R injury. Sections at ×200 magnification. Scale bars represent 100 μm. Experiments were performed with n = 6. Graphs represent mean ± SEM. Statistical significance is marked as * p < 0.05, ** p < 0.01, ***p > 0.001 (One-way ANOVA with Bonferroni's Multiple Comparison post-test).
Cellular location of TWEAK
For further evidence of TWEAK expression levels, we performed immunostaining on muscle sections of healthy Nrf2-WT and KO mice as well as on muscle sections of injured mice at 6 h, 96 h and 14 days after ischemia (Fig. 1 C–J). Although no major differences between Nrf2-WT and KO mice could be observed in uninjured animals, Nrf2-KO mice show distinct TWEAK-positive membranes (Fig. 1 D). 6 h after ischemia, one can see few TWEAK-positive nuclei in the myofibres, but otherwise no positive signal (Fig. 1E, F).
96 h after ischemia, cells have become highly TWEAK-positive, with the Nrf2-KO mice showing much more intense staining than the Nrf2-WT mice (Fig. 1G, H). Skeletal muscle cells have been replaced by myotubes, the precursors of mature myofibres. These have central, large nuclei that contrast with the small, peripheral nuclei of mature myofibres. The positive signal is located both in the sarcoplasm and in nuclei.
14 days after ischemia Nrf2-WT mice have almost regained normal physiological-morphological structure and have become mostly TWEAK-negative again. Nrf2-KO mice still show TWEAK-positive, immature myofibres of varying, irregular calibre (Fig. 1I, J).
Nrf2-KO mice show an increased number of apoptotic cells after I/R injury
To evaluate the proportion of dead cells we performed a TUNEL assay that labels apoptotic nuclei (Fig. 2). Neither the healthy muscle of Nrf2-WT or KO mice nor either group's muscle 96 h after ischemia contains apoptotic nuclei (see supplementary Fig. S1). In contrast apoptotic nuclei are present in high proportion 6 h after ischemia, with Nrf2-WT mice showing a considerably smaller portion of apoptotic nuclei (40.67 ± 4.82%) as compared to Nrf2-KO mice (66 ± 4.95%) (Fig. 2G).
Nrf2-KO mice show more apoptotic cells after I/R injury.
(A) TUNEL Assay was performed to detect apoptotic nuclei on sections Nrf2-WT (A–C) and KO mice (D–F) 6 h after I/R injury. The number of apoptotic nuclei was manually determined and calculated as a ratio of the total cell number (G). Sections at ×200 magnification. Scale bar represent 100 μm. Experiments were performed with n = 6. Graphs represent mean ± SEM. Statistical significance is marked as * p < 0.05, ** p < 0.01 (Unpaired Two-tailed Student's t-test).
Nrf2-KO mice show enhanced fibrosis and insufficient regeneration
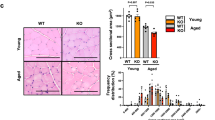
We performed Ladewig and Sirius-red staining on muscle sections from all time groups in order to characterize possible differences in the respective morphological structures of Nrf2-WT and KO mice (Fig. 3). Ladewig staining is suitable for revealing tissue fibrosis, since it causes red colouration in mature myofibres but blue colouration in fibrotic tissue. Sirius-red staining causes red colouration of collagen fibres and was evaluated by polarised light microscopy. These techniques enable sensitive discrimination of the extent of fibrosis.
Nrf2-KO mice exhibit extended fibrosis and insufficient regeneration after I/R injury.
Ladewig staining (A–H) and Sirius-red staining (I–P) was performed on sections of healthy Nrf2-WT and KO mice (A,B/I,J) each at 6 h (C,D/K,L), 96 h (E,F/M,N) and 14 d (G,H/O,P) after I/R injury. Fibrotic tissue is stained blue in Ladewig staining (black arrows) and red in Sirius-red staining (white arrows). Sirius-red staining was examined under polarised light. Sections at ×200 magnification.
Neither healthy Nrf2-WT nor KO mice show any fibrosis (Fig. 3A,B/I,J), nor does either group 6 h after ischemia. At the 6 h interval infiltration by neutrophil granulocytes has taken place and cell connections between myotubes have been dissolved (Fig. 3C, D). 96 h after ischemia, however, macrophages have infiltrated the injured muscle. Myofibres have been replaced by so-called myotubes, with their characteristic central nuclei. The basophilic staining hints at increased amounts of mRNA and ribosomes from increased protein production (Fig. 3E, F). Stripes of connective tissue are visible between the myotubes (black arrows in Fig. 3E, F/white arrows in M,N). Nrf2-KO mice are far more interspersed with these fibrotic stripes than are the Nrf2-WT mice. 14 d after ischemia, Nrf2-WT mice have nearly fully recovered from injury; they show integer architecture with few central nuclei that have not yet moved to the periphery (Fig. 3G). In contrast Nrf2-KO mice still show traces of infiltration, huge calibre variations and remaining fibrosis (black arrows in Fig. 3H/white arrows in Fig. 3P).
TWEAK activates Nrf2 in C2C12 cells
In order to assess the effect of TWEAK on Nrf2-acitivty we established a luciferase reporter assay by which we measured Nrf2-dependent ARE-activity. For this we used a C2C12 cell line with stable transfection of pGL4-ARE. C2C12-ARE-transfected cells were deprived of growth factors in FCS-free medium to initiate differentiation and form myotubes. Differentiated cells were stimulated for 6 h, 12 h or 24 h, each at concentrations of 30 ng/ml (yellow bars) and 60 ng/ml (blue bars) of TWEAK (Fig. 4A). After 6 h stimulation at either concentration, no significant increase in ARE activity appears as compared to unstimulated control. After 12 h stimulation, however, the 60 ng/ml TWEAK concentration has produced twice as much ARE activity as could be observed in unstimulated control, a significant upregulation. After 24 h even a 30 ng/ml-concentration of TWEAK has produced a doubling in upregulation of ARE-activity, while 60 ng/ml of TWEAK has increased ARE activity by approximately three times. To elucidate the signalling pathway by which TWEAK stimulation activates Nrf2, C2C12 cells were pre-treated with various kinase inhibitors (see supplements, Table S1). Inhibition of p38, ERK and JNK abolished the observed TWEAK-mediated Nrf2 activation (Fig. 4B, fourth, fifth and seventh bar). In contrast, inhibition of PI3K, GRK-5 and PKC did not prevent Nrf2 activation by TWEAK.
TWEAK activates Nrf2 and Nrf2 protects against TWEAK-mediated cell death.
(A) C2C12 cells were stimulated with 30 (yellow bars) or 60 ng/ml (blue bars) of TWEAK for 6 h, 12 h or 24 h. (B) 30 minutes prior to stimulation with 60 ng/ml of TWEAK for 24 h, kinase inhibitors for p38 (SB = SB 203580), ERK (PD = PD 98059), PI3K (Wo = Wortmannin), JNK (SP = SP 600125), GRK-5 and PKC (Ro = Ro-31-8220). (C) A cytotoxicity assay was performed with C2C12 cells transfected with shRNA against Nrf2 (red) or Keap1 (green) and non-target control shRNA (grey), all stimulated with 30, 60 or 120 ng/ml of TWEAK for 24 h. (D) C2C12 cells with non-target shRNA (grey bars), shRNA against Nrf2 (red bars) and Keap1 (green bars) were transfected with an NFkB-binding site-luciferase construct. Cells were stimulated with 60 ng/ml of TWEAK for 12 h. Additionally, C2C12 cells were co-stimulated with 60 ng/ml of TWEAK and 150 μM of Methysticin (blue bar). (E) Western blot analysis with muscle samples from Nrf2-WT and KO mice 6 h (left lane) and 96 h (right lane) after I/R was used to compare the expression of NFkB (65 kDa). GAPDH was used as an internal loading control. The full-length blots with anti-NFkB and anti-GAPDH antibodies are presented in supplementary Figure S2. All gels have been run simultaneously under the same experimental conditions. Cell culture experiments were performed at n = 8, in vivo experiments at n = 6. Graphs represent mean ± SEM. Statistical significance is marked as * p < 0.05, ** p < 0.01, ***p < 0.001 (One-way ANOVA with Bonferroni's Multiple Comparison post-test).
Nrf2-knockdown augments cell death
It is well established that TWEAK leads to skeletal muscle wasting and cell death12. To see whether Nrf2 deficiency might impact differentiated myotubes, we established stable knockdowns of Nrf2 and Keap1 in C2C12 cells. These cells were stimulated with 30, 60 or 120 ng/ml of TWEAK each for 24 h (Fig. 4C). Cell death did not emerge in control C2C12 myotubes (grey bars) until treated with TWEAK at 120 ng/ml. In contrast, Nrf2-knockdown C2C12 cells (red bars) already exhibited sensitivity to TWEAK at a concentration of 30 ng/ml and were also sensitive at the higher concentrations. In contrast, Nrf2-knockdown C2C12 cells (red bars) already exhibited sensitivity to TWEAK at a concentration of 30 ng/ml and were also sensitive at the higher concentrations. Keap1-knockdown C2C12 cells (green bars), which over-express Nrf2, exhibited almost the same characteristics as normal C2C12 cells except that they showed resistance to 120 ng/ml of TWEAK. Thus the extent of cell death does not increase in C2C12 cells that lack Keap1.
TWEAK repression by Nrf2 is mediated via NFkB activity
The interaction between Nrf2 and NFkB has been already investigated in the past6. NFkB activity was measured in TWEAK-treated C2C12 cells with Nrf2- and Keap1-knockdwon (Fig. 4D). TWEAK induced NFkB activity in control C2C12 cells (third bar, grey), an effect that was further increased by knockdown of Nrf2 (fourth bar, red). In contrast, knockdown of Keap1 (fifth bar, green), which leads to Nrf2 activation, as well as Methystcin induced Nrf2 activation13 (last bar, orange) significantly reduced NFkB activity. Moreover, NFkB expression was measured in Nrf2-WT and -KO mice via Western blotting 6 h (Fig. 4E, left lane) and 96 h after I/R injury (Fig. 4E, right lane). At both time points, Nrf2-KO mice showed a significantly greater expression of NFkB as compared to Nrf2-WT mice.
Discussion
Since TWEAK has been described as a very potent inducer of skeletal muscle wasting12 and skeletal muscle fibrosis4, we aimed to investigate the status of TWEAK expression in Nrf2-deficient mice and how this might affect the outcome of skeletal muscle I/R injury and regeneration.
In contrast to wild type mice, Nrf2-deficient mice did significantly upregulate TWEAK expression after I/R injury (Fig. 1). These results are in line with other findings on pro-inflammatory cytokine expression in Nrf2-KO mice. Ichihara et al. showed that Nrf2-KO mice express higher amounts of TNFalpha and IL-68. Also, Thimmulappa et al. showed that Nrf2-KO mice have greater activation of TNFalpha and NF-kappaB in response to LPS treatment9. Khor et al. further confirmed the increased expression of pro-inflammatory cytokines in inflammatory bowel disease of Nrf2-KO mice10. We therefore can add TWEAK to this group of cytokines that are influenced by Nrf2.
Little is known about the intracellular localization of TWEAK in skeletal muscles. Immunofluorescence with anti-TWEAK-antibodies located it in the sarcoplasm (Fig. 1H) as well as in the nuclei (Fig. 1J). Since TWEAK is thought to be a transmembrane protein14, it is endogenously produced in skeletal muscle cells as well as in infiltrating macrophages. An explanation for TWEAK being found in the sarcoplasm is that skeletal muscle cells may have stores of TWEAK ready to be released in the event of stress or injury. In fact, one recent study on inflammatory bowel disease found TWEAK to be localized in cytoplasm in colonocytes15.
Moreover, we also found that regenerated myofibres have TWEAK-positive nuclei (Fig. 1J). This is interesting because it has been reported that TWEAK contains nuclear localization sequences16,17. Our data confirm for the first time that these nuclear localization sequences are indeed functional, because the nuclei in the muscles after injury are TWEAK-positive. The functional role of TWEAK in the nucleus, however, remains to be elucidated.
One consequence of I/R injury is the induction of muscle cell apoptosis. A TUNEL assay showed a significantly higher number of apoptotic nuclei in Nrf2-KO mice (Fig. 2), which is in line with the higher levels of TWEAK18. We confirmed this finding in vitro utilising C2C12 cells (Fig. 4), which proves that Nrf2 protects against TWEAK-mediated cell death. The decision on cell survival or death is regulated by p53, which, in case of high levels of stress, acts in a proapoptotic fashion by inhibiting Nrf2 and activating multiple genes to induce full commitment to cell death19. It has already been shown that Nrf2 can suppress p53 expression under certain circumstances20. This may explain why C2C12 cells with overexpressed Nrf2 show higher resistance to TWEAK concentrations and maintain higher viability.
We show here that the mechanism behind the Nrf2-induced suppression of TWEAK depends on NFkB activity. NFkB is a main regulator of inflammation and orchestrates different steps of inflammatory reactions. Recent studies have shown how Nrf2 activity can dampen inflammation, particularly through downregulation of the pro-inflammatory factor NFkB21,22. Since TWEAK expression is upregulated by NFkB via a functional NFkB binding site in its promoter16, we suggest that this might be an intercept point between the Nrf2 and NFkB pathway - Nrf2 attenuates NFkB expression and activity in Nrf2 activated Keap-1 knockdown and methysticin treated C2C12 cells (Fig. 4D). Moreover, Nrf2-KO mice showed higher expression levels of NFkB as compared to Nrf2-WT mice 6 h and 96 h after I/R (Fig. 4E). Therefore, we conclude that the suppression of TWEAK expression by Nrf2 is mediated via NFkB inhibition (Fig. 5).
Suggested mechanism for TWEAK-Nrf2 crosstalk.
TWEAK binds to its receptor and activates an intracellular signalling cascade (*)12,23, which leads to phosphorylation of Keap1 or accordingly Nrf2 via various mitogen activated kinases27. This induces a disinhibition of Nrf2, which then represses NFkB expression as shown elsewhere (#)6,22. Its inhibition consequently leads to downregulation of TWEAK, as NFkB normally induces TWEAK expression (§)16.
Interestingly, TWEAK is a potent inducer of Nrf2 (Fig. 4A) and therefore reduces its own expression in a negative feedback loop. It has been shown that TWEAK signalling depends on various mitogen activated kinases like p38, ERK or JNK12,23; by inhibition of these kinases, the TWEAK-induced Nrf2 activation was abolished, pointing out that Nrf2 is a link in the chain of TWEAK signalling. This is in line with earlier publications that showed that kinases can activate Nrf224,25,26,27.
Several studies have shown that skeletal muscle wasting and severe interstitial fibrosis come about in TWEAK-transgenic mice or in mice treated with recombinant TWEAK12,28. Furthermore, ablation of TWEAK improves the outcome of post-injury muscle regeneration in mice4. In order to evaluate tissue regeneration and fibrosis, we performed Ladewig and Sirius-red staining (Fig. 3). The staining showed that Nrf2-deficient mice show high amounts of interstitial and intercellular fibrosis, whereas Nrf2-WT mice only exhibit very limited traces of fibrotic tissue (Fig. 3E, F). This discrepancy between the two genotypes is maintained until 14 days, at which point NRf2-WT mice have fully recovered from injury, while Nrf2-KO mice still show immature myofibres with central nuclei, traces of inflammatory cells and, most notably, fibrotic tissue (Fig. 3G, H). Therefore, we can conclude that the outcome of injury to Nrf2-KO mice is comparable to that found in TWEAK-transgenic mice.
In conclusion, the results of the current study show that skeletal muscle of mice depleted of Nrf2 have elevated levels of TWEAK after I/R injury. This overproduction of TWEAK may be responsible for the insufficient regeneration of muscle deficient in Nrf2. Furthermore, we show that TWEAK stimulation activates the transcription factor Nrf2 (Fig. 4A), while Nrf2 activity protects against the atrophic effect of TWEAK (Fig. 4B). Taken together, the understanding of mechanisms that conserve the skeletal muscle may lay the ground for novel therapeutic strategies for the treatment of skeletal muscle wasting and atrophy.
Methods
Animal trials
All experiments were conducted in accordance with German legislation governing animal studies. The Principles of Laboratory Animal Care were followed. The animal experiments were approved by the “Landesamt fuer Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen” (Reg. 8.87–50.10.37.09.209).
To ensure a sterile injury, skeletal muscle I/R was induced by applying a tourniquet around one hindlimb for 4 hours29. Muscle samples were harvested after 6 h, 96 h or 14 d and were formalin-fixed for histology or shock-frozen for homogenization.
Animals
C57BL/6 mice were kept in our animal facility under pathogen-free conditions on a 12-hour night, 12-hour light schedule. Mice were given water and fed with standard rodent food ad libitum. All mice used for this study were between 10 and 12 weeks old. Nrf2-Knock-out mice were obtained by specific deletion of the Nrf2 gene segment30.
Luciferase assay
After stimulation, transfected cells were washed with sterile PBS. Cells were lysed by adding 100 μl of passive lysis buffer (Promega, Madison, WI) to each well of a 96-well plate. 50 μl of the lysate were transferred to a 96-well microtiter plate for luminescence measurement in a GLOMAX luminometer (Promega). Luminescence was measured over 10 s using a commercially available Luciferase Reporter Assay (Promega). For quantification, 50 μl of the remaining lysate were used to determine cell number with CyQuant Cell Proliferation Assay (Invitrogen). Data were analysed by normalizing luminescence signals to the corresponding CyQuant signal of each well.
Cell culture and transfection
Murine C2C12 myoblast cells were grown in standard DMEM medium plus 10% foetal calf serum and 1% penicillin-streptomycin. Cells were seeded in 96-well plates containing 100 μl medium and approx. 1 × 104 cells per well. For stable transfection, shRNA against Nrf2 and Keap1 was transduced into C2C12 cells via lentiviruses and selection was achieved with 2.5 mg/ml of puromycine in standard DMEM medium.
For the NFkB reporter assay, an NFkB-activity reporter construct that contained an NFkB binding site together with a luciferase gene was transfected into C2C12 cells via lentiviruses.
Stimulation and differentiation treatment
For observation and stimulation of fully-differentiated myofibres, cells were grown to full confluency and subsequently treated with low-serum differentiation medium, which included 1% FCS, until myotubes were visible31. Cells were then submitted to FCS-free medium containing 30, 60 and 120 ng/ml respectively of recombinant TWEAK (R&D Systems, #1237-TW-025). The stimulation was sustained for 24 h.
Signalling inhibition
Specific pathway inhibitors were used to stop signal transduction of TWEAK (see supplements, Table S1).These substances were applied to the cells 30 minutes before TWEAK stimulation. Nrf2-activity was measured after 24 h hours via luciferase assay (see above).
Cytotoxicity assay
The Cytotox-Glo cytotoxicity assay (Promega) was used to measure cytotoxicity. Differentiated C2C12 cells were stimulated with 30, 60 or 120 ng/ml of TWEAK for 24 h; the kit was used according to the manufacture's protocol. Luminescence was detected in a 96-well plate reader (Glomax microplate luminometer, Promega).
mRNA-expression
RNA was extracted from muscle homogenates using peqGOLD TriFast (Peqlab Biotechnology GmbH) according to the manufacturer's protocol. Transcription into cDNA was done using a VILO cDNA Synthesis kit (Invitrogen). RT-PCRs were performed using the StepOnePlus System (Applied Biosystems); reagant mixtures were transferred to a 96-well plate, each well containing 20 μl of 100 ng cDNA, primers and SYBR Green I reagent (Applied Biosystems). To determine gene expression, target-gene Ct values were normalized to the expression of the housekeeping gene succinate dehydrogenase A (SDHA2). The sequences for the SDHA2primer (Eurofins MWG Operon) were 5′-AGGTCGGTGT-GAACGGATTTG-3′ (forward) and 5′-TGTAGACCATGTAGTTGAGGTCA-3′ (reverse) with an annealing temperature of 60.5°C; those for the TWEAK primer (Eurofins MWG Operon) were 5′-GGAACACTCCAAAAACAGACCT-3′ (forward) and 5′- CCACCACTGGGTATTGAGTAGAA-3′ (reverse) with an annealing temperature of 60°C.
ELISA
Muscle samples were harvested 96 h and 14 days after hindlimb I/R and from healthy control mice. 1 ml of ice-cold Lysis Buffer A (Abcam Plc, Cambridge, UK) was added to approx. 30 mg of tissue and probes were homogenized in an electric homogenizer (Precellys 24, Peqlab). Protein concentration was measured with a BCA Protein Assay according to the manufacturer's protocol (Thermo Fisher Scientific Inc., IL, USA). TWEAK levels were measured by a sandwich ELISA (R&D Systems).
Histology
Muscle samples were fixed in 3.5% formaldehyde, embedded in paraffin and cut into 5 μm-thick sections. Sections were stained with an antibody against TWEAK (sc-5558, 1:100, SantaCruz Biotechnology). Bound primary antibodies were detected by immunofluorescence using secondary antibodies conjugated with Cyanine5 (Jackson ImmunoResearch Laboratories). Nuclei were stained with Bisbenzimid (Hoechst 33258). Control sections were treated similarly, except for the primary antibody, which was omitted.
For Ladewig staining, sections were brought to water with xylene and ethanol and treated with Ladewig's and Weigert's solution. For Sirius-red staining, sections were treated with Sirius-red solution (#365548, Sigma-Aldrich) for one hour and washed with acetic acid.
Digital evaluation and quantification were done on ImageJ software (National Institutes of Health, Bethesda, MD, USA.)
Western blot analysis
Proteins were subjected to 12.5% discontinuous SDS-PAGE, separated by electrophoresis and electroblotted to PVDF membranes (Milipore Corp., Billerica, MA, USA). Specific antibodies against NFkB (#3033, 1:1000 in 5%-BSA-TBST, Cell Signalling) were used to detect bands at 65 kDa. For the corresponding loading control, an antibody against GAPDH was used (gt239, 1:2000, Genetex Inc.). Primary antibodies were incubated at 4°C over night, the according HRP-Secondary antibodies were incubated for one hour at room temperature and binding was detected with a chemiluminescence-based detection kit (Immobilon Western, Milipore Corp.).
Statistical analysis
Statistical analyses were performed using Student's unpaired t-test for dual comparisons or one-way analysis of variance for multiple comparisons. Mean differences were considered significant at p < 0.05. Regression calculations were done with Excel 2007 (Microsoft Corporation, Redmond, WA). GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA) was used for statistical graphs and analyses.
References
Sharma, R. & Anker, S. D. Cytokines, apoptosis and cachexia: the potential for TNF antagonism. Int J Cardiol 85, 161–171 (2002).
Bhatnagar, S., Mittal, A., Gupta, S. K. & Kumar, A. TWEAK causes myotube atrophy through coordinated activation of ubiquitin-proteasome system, autophagy and caspases. J Cell Physiol 227, 1042–1051, 10.1002/jcp.22821 (2012).
Dogra, C. et al. TNF-related weak inducer of apoptosis (TWEAK) is a potent skeletal muscle-wasting cytokine. FASEB J 21, 1857–1869, 10.1096/fj.06-7537com (2007).
Mittal, A. et al. Genetic ablation of TWEAK augments regeneration and post-injury growth of skeletal muscle in mice. Am J Pathol 177, 1732–1742, 10.2353/ajpath.2010.100335 (2010).
Wasserman, W. W. & Fahl, W. E. Functional antioxidant responsive elements. Proc Natl Acad Sci U S A 94, 5361–5366 (1997).
Wakabayashi, N., Slocum, S. L., Skoko, J. J., Shin, S. & Kensler, T. W. When NRF2 talks, who's listening? Antioxid Redox Signal 13, 1649–1663, 10.1089/ars.2010.3216 (2010).
Kim, J., Cha, Y. N. & Surh, Y. J. A protective role of nuclear factor-erythroid 2-related factor-2 (Nrf2) in inflammatory disorders. Mutat Res 690, 12–23, 10.1016/j.mrfmmm.2009.09.007 (2010).
Ichihara, S. et al. Ablation of the transcription factor Nrf2 promotes ischemia-induced neovascularization by enhancing the inflammatory response. Arterioscler Thromb Vasc Biol 30, 1553–1561, 10.1161/ATVBAHA.110.204123 (2010).
Thimmulappa, R. K. et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J Clin Invest 116, 984–995, 10.1172/jci25790 (2006).
Khor, T. O. et al. Nrf2-deficient mice have an increased susceptibility to dextran sulfate sodium-induced colitis. Cancer Res 66, 11580–11584, 10.1158/0008-5472.can-06-3562 (2006).
Osburn, W. O. et al. Increased colonic inflammatory injury and formation of aberrant crypt foci in Nrf2-deficient mice upon dextran sulfate treatment. Int J Cancer 121, 1883–1891, 10.1002/ijc.22943 (2007).
Bhatnagar, S. & Kumar, A. The TWEAK-Fn14 system: breaking the silence of cytokine-induced skeletal muscle wasting. Curr Mol Med 12, 3–13 (2012).
Wruck, C. J. et al. Kavalactones protect neural cells against amyloid beta peptide-induced neurotoxicity via extracellular signal-regulated kinase 1/2-dependent nuclear factor erythroid 2-related factor 2 activation. Mol Pharmacol 73, 1785–1795, 10.1124/mol.107.042499 (2008).
Winkles, J. A. The TWEAK-Fn14 cytokine-receptor axis: discovery, biology and therapeutic targeting. Nat Rev Drug Discov 7, 411–425, 10.1038/nrd2488 (2008).
Dohi, T., Kawashima, R. & Burkly, L. C. Role of TWEAK (TNF-alpha-Like Weak Inducer of Apoptosis) in Intestinal Inflammation and Tissue Repair. Gastroenterology 136, A255–A255 (2009).
Baxter, F. O. et al. IKKbeta/2 induces TWEAK and apoptosis in mammary epithelial cells. Development 133, 3485–3494, 10.1242/dev.02502 (2006).
De Ketelaere, A. et al. Involvement of GSK-3beta in TWEAK-mediated NF-kappaB activation. FEBS Lett 566, 60–64, 10.1016/j.febslet.2004.04.041 (2004).
Chicheportiche, Y. et al. TWEAK, a new secreted ligand in the tumor necrosis factor family that weakly induces apoptosis. Journal of Biological Chemistry 272, 32401–32410, 10.1074/jbc.272.51.32401 (1997).
Faraonio, R. et al. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem 281, 39776–39784, 10.1074/jbc.M605707200 (2006).
Kwak, M. K. et al. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem 278, 8135–8145, 10.1074/jbc.M211898200 (2003).
Li, W. et al. Activation of Nrf2-antioxidant signaling attenuates NFkappaB-inflammatory response and elicits apoptosis. Biochem Pharmacol 76, 1485–1489, 10.1016/j.bcp.2008.07.017 (2008).
Folmer, F. et al. Inhibition of TNFalpha-induced activation of nuclear factor kappaB by kava (Piper methysticum) derivatives. Biochem Pharmacol 71, 1206–1218, 10.1016/j.bcp.2005.12.032 (2006).
Li, H. et al. Tumor necrosis factor-related weak inducer of apoptosis augments matrix metalloproteinase 9 (MMP-9) production in skeletal muscle through the activation of nuclear factor-kappaB-inducing kinase and p38 mitogen-activated protein kinase: a potential role of MMP-9 in myopathy. J Biol Chem 284, 4439–4450, 10.1074/jbc.M805546200 (2009).
Yu, R. et al. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J Biol Chem 275, 39907–39913, 10.1074/jbc.M004037200 (2000).
Kweider, N. et al. Interplay between vascular endothelial growth factor (VEGF) and nuclear factor erythroid 2-related factor-2 (Nrf2): implications for preeclampsia. J Biol Chem 286, 42863–42872, 10.1074/jbc.M111.286880 (2011).
Wruck, C. J. et al. Luteolin protects rat PC12 and C6 cells against MPP+ induced toxicity via an ERK dependent Keap1-Nrf2-ARE pathway. J Neural Transm Suppl 57–67 (2007).
Bryan, H. K., Olayanju, A., Goldring, C. E. & Park, B. K. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol 85, 705–717, 10.1016/j.bcp.2012.11.016 (2013).
Mittal, A. et al. The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice. J Cell Biol 188, 833–849, 10.1083/jcb.200909117 (2010).
Sönmez, T. T. et al. A Novel Laser-Doppler Flowmetry Assisted Murine Model of Acute Hindlimb Ischemia-Reperfusion for Free Flap Research. PLOS ONE 8, e66498 (2013).
Chan, K., Lu, R., Chang, J. C. & Kan, Y. W. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth and development. Proc Natl Acad Sci U S A 93, 13943–13948 (1996).
Jahn, L., Sadoshima, J. & Izumo, S. Cyclins and cyclin-dependent kinases are differentially regulated during terminal differentiation of C2C12 muscle cells. Exp Cell Res 212, 297–307 (1994).
Acknowledgements
We acknowledge funding from the START program of the Faculty of Medicine of RWTH Aachen University. This study was supported in part by the Deutsche Forschungsgemeinschaft (DFG) (DFG No. PU 214/3-2: 4-2; 5-2) and by a grant from the Interdisciplinary Centre for Clinical Research (IZKF) at the Faculty of Medicine at the RWTH Aachen University (T9-3; T9-5).
Author information
Authors and Affiliations
Contributions
This study was conceived and designed by O.A. and C.J.W. O.A. was the primary experimentalist. Y.W.K. generated the Nrf2 knockout mice. A.F. and C.R. provided shRNA-constructs, critical advice on experimental procedures and data analysis. T.P. and T.T.S. supported the execution of this study. The manuscript was written by O.A. and C.J.W.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Al-Sawaf, O., Fragoulis, A., Rosen, C. et al. Nrf2 Protects Against TWEAK-mediated Skeletal Muscle Wasting. Sci Rep 4, 3625 (2014). https://doi.org/10.1038/srep03625
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03625
This article is cited by
-
Transcription factor NRF2 as potential therapeutic target for preventing muscle wasting in aging chronic kidney disease patients
Journal of Nephrology (2022)
-
Role of Nrf2 in Fracture Healing: Clinical Aspects of Oxidative Stress
Calcified Tissue International (2019)
-
Partial involvement of Nrf2 in skeletal muscle mitohormesis as an adaptive response to mitochondrial uncoupling
Scientific Reports (2018)
-
The role of oxidative stress in skeletal muscle injury and regeneration: focus on antioxidant enzymes
Journal of Muscle Research and Cell Motility (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.