Abstract
Next-generation sequencing (NGS) is revolutionising marker development and the rapidly increasing amount of transcriptomes published across a wide variety of taxa is providing valuable sequence databases for the identification of genetic markers without the need to generate new sequences. Microsatellites are still the most important source of polymorphic markers in ecology and evolution. Motivated by our long-term interest in the adaptive radiation of a non-model species complex of whitefishes (Coregonus spp.), in this study, we focus on microsatellite characterisation and multiplex optimisation using transcriptome sequences generated by Illumina® and Roche-454, as well as online databases of Expressed Sequence Tags (EST) for the study of whitefish evolution and demographic history. We identified and optimised 40 polymorphic loci in multiplex PCR reactions and validated the robustness of our analyses by testing several population genetics and phylogeographic predictions using 494 fish from five lakes and 2 distinct ecotypes.
Similar content being viewed by others
Introduction
In the early 1980's, the discovery of simple-sequence repeats (SSR), or microsatellites, seemed to be insignificant, given the laborious cloning and sequencing procedures that were required to analyse the polymorphisms1. However during the last two decades, they have arguably become the most important and versatile source of polymorphic genetic markers for the construction of linkage maps, parentage testing, population and conservation genetics, management of biological resources and other related fields2. Microsatellite-generated distances have also proven to be powerful estimators of recent demographic events3 and empirical tests of their reliability for constructing phylogenetic trees have led to the development of appropriate methodological approaches to perform these types of analyses at different evolutionary scales4. Until recently, the major obstacle for using microsatellite markers for population studies was the slow and costly de novo isolation and characterisation of a set of polymorphic loci in the focal species. However, next-generation sequencing (NGS) and improved bioinformatics tools are revolutionising microsatellite marker development5,6. Furthermore, the rapidly increasing amount of transcriptomes and genomes published across a wide variety of taxa provides valuable sequence databases for the identification of SSR without the need to generate new sequences7. Given the presence of such databases for a species of interest, this approach provides a cost-effective way for research projects working on modest financial resources to identify a suitable starting set of microsatellites at virtually no laboratory costs. Furthermore, even when such databases are not available, the current costs of NGS for microsatellite detection across taxa are only a fraction of what it used to be using traditional methods6.
Motivated by our long-term interest in the adaptive radiation of a non-model species complex of whitefishes (Coregonus spp.) reviewed in8, in this study, we focus on microsatellite characterisation and multiplex optimisation of a large number of loci in Coregonus clupeaformis (Mitchill, 1818) using transcriptome sequences generated by two NGS platforms (Illumina® and Roche-454), as well as online databases of Expressed Sequence Tags (EST). Furthermore, we assessed the genotyping robustness of the characterised loci in assessing the evolutionary relationships of the species complex by constructing population phenograms of previously studied sympatric ecotypes from different lakes. Lake whitefishes inhabit hundreds of postglacial freshwater bodies in the northern hemisphere, which harbour pairs of divergent whitefish morphs or ecotypes that have been evolving during the past 15,000 years9,10. They are characterised by the presence of two sympatric ecotypes with different morphology and ecology. The benthic ecotype is the ancestral form and is called "normal", while the limnetic ecotype is much smaller and elongated and is known as "dwarf". These two ecotypes have evolved independently and repeatedly in at least six different lakes of the Saint-John River watershed in the last 12,000 years11. These ecotype pairs display parallelism in life history, behavioural and morphological divergence associated with the use of distinct trophic resources8,12. The extent of genetic differentiation within sympatric ecotype pairs varies substantially among different lakes. This differentiation can range from extensive introgression to complete reproductive isolation, depending on historical contingencies13,14,15 and also on the potential for competitive interactions imposed by the unique ecological characteristics of each lake14,16,17. Lake whitefishes thus represent a rarely paralleled example of taxa displaying a continuum of both morphological and genetic differentiation, making it an ideal model species for the study of speciation and parallel evolution.
Our primary goal was to rapidly generate molecular resources by obtaining a sufficient number of microsatellites for future studies on the evolution and demographic history of lake whitefishes. We wanted to develop an approach for rapidly and inexpensively identifying informative, polymorphic microsatellite loci by leveraging available high-throughput sequencing databases, screening for amplification and variation and then scoring the most promising loci. Furthermore, we also aimed at comparing two NGS platforms to generate molecular resources for a non-model species. Here, we identified thousands of SSR loci with flanking sequences suitable for the design of PCR primers and optimised 39 loci into 8 multiplex panels, permitting the PCR amplification of several markers simultaneously and tested the robustness of our analyses by confirming phylogenetic predictions set from several studies that have documented the adaptive divergence of this species complex. These multiplexes drastically reduce laboratory costs and labour time, enable high-throughput analyses and promote accuracy as well as precision of the genetic results18.
Results
Microsatellite loci in EST
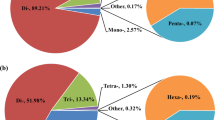
From the 627,608 reads generated by Roche-454 sequencing and assembled into 14,449 contigs, as reported in Renaut, et al.19, we identified 2,267 contigs (15.7%) that contained microsatellite loci with di- to octa-nucleotide repetitions. Of these, there was a total of 349, 157 and 61 contigs that contained Potentially Amplifiable Loci (PAL) of di- tri- and tetra-nucleotide repeats respectively and 369 non-PAL totalling 936 contigs with these classes of SSR. From the 6,727 cDNA contigs available online20, we identified 1,391 contigs (20.5%) that contained microsatellite loci with di- to octa-nucleotide repetitions. Of these, there was a total of 266, 124 and 55 contigs that contained di- tri- and tetra-nucleotide repeats respectively, totalling 549 contigs, of which we identified 445 PAL. Finally, the 628,416,550 paired-end (PE) reads generated by Illumina® HiSeq™ 2000 Sequencing Systems were assembled into 75,905 contigs (AM Dion-Côté and L Bernatchez, unpublished), where we identified 28,784 contigs (37.9%) that contained microsatellite loci with di- to octa-nucleotide repetitions. Of these, there was a total of 5,521, 3,534 and 939 contigs that contained di- tri- and tetra-nucleotide repeats respectively, totalling 12,156 contigs, of which we identified 9,994 PAL (Table 1, Figure 1). In total, we thus identified 32,442 EST contigs containing microsatellites, which represented 33.4% of all contigs, of which 33.9% were identified as PAL of di- tri- and tetra-nucleotide microsatellites and 8.1% as non-PAL (i.e. EST contigs containing di- tri- and tetra-nucleotide microsatellites without suitable flanking PCR-primer sites). Comparing across the three classes of repeats, di-nucleotide repeats represented 55.7% (6,136), tri-nucleotides 34.7%, (3,815) and tetra-nucleotides 9.6% (1,055) of the potentially amplifiable loci, respectively (Table 1, Figure 2). Finally, out of the 937 sequences from the Roche-454 dataset containing PAL with SSR, 386 were also found among the 9244 sequences from the Illumina dataset (blastn options: -evalue 1e-10 -perc_identity 90). Among the 44 PAL with SSR loci developed, only 5 were found among the 386 loci present in both datasets.
Evaluation of the number of microsatellite-containing EST from the different datasets.
The first step included all potential loci from the initial transcriptome contigs, the second step filtered those and included loci containing di- to octa-nucleotides, the third step included loci containing di- tri- and tetra-nucleotides and the fourth step included loci useful for primer design for successful PCR amplification (i.e. with sufficiently large flanking sequences around the microsatellite).
Microsatellite development
From the EST containing microsatellites, we selected 44 microsatellite loci for testing their usefulness in multiplex genotyping and 38 produced PCR amplification when visualized on agarose gels and ethidium bromide staining. Of these 44 loci, 9 gave multiple or faint bands or fragments clearly defined but of a different size than the one expected from the original sequence. These loci were discarded from subsequent analyses. Of the remaining 29, 22 could be easily genotyped and displayed polymorphism in 48 normal fish from all lakes (Supplementary Table 1). One locus, Cocl11, amplified 2 unlinked loci. The number of alleles per locus ranged from 2 to 25 ( , σ = 6.26). Expected and observed heterozygosities ranged from 0.05 to 0.83 (
, σ = 6.26). Expected and observed heterozygosities ranged from 0.05 to 0.83 ( , σ = 0.20) and from 0.05 to 0.88 (
, σ = 0.20) and from 0.05 to 0.88 ( , σ = 0.23), respectively. There was no evidence of large allele dropout or genotyping errors due to stutter peaks for 20 loci. However, 2 loci (Cocl14 and Cocl28) showed a significant excess of homozygotes (deviation from Hardy– Weinberg equilibrium, P < 0.001; Supplementary Table 1). There was evidence of linkage disequilibrium for only one pair of loci (Cocl11 and Cocl20) (P < 0.001). Genotyping was consistent across the 48 samples, which were tested twice and yielded the same genotypes. Although all these loci are located in transcribed sequences, BLAST searches uncovered links to protein sequences in only 3 of these 22 microsatellite sequences. Locus Cocl17 matched with a Follistatin-related precursor in Salmo salar, locus Cocl40 matched with a NEDD4-binding like protein and locus Cocl44 matched with an ELAV-like protein 1 in Salmo salar.
, σ = 0.23), respectively. There was no evidence of large allele dropout or genotyping errors due to stutter peaks for 20 loci. However, 2 loci (Cocl14 and Cocl28) showed a significant excess of homozygotes (deviation from Hardy– Weinberg equilibrium, P < 0.001; Supplementary Table 1). There was evidence of linkage disequilibrium for only one pair of loci (Cocl11 and Cocl20) (P < 0.001). Genotyping was consistent across the 48 samples, which were tested twice and yielded the same genotypes. Although all these loci are located in transcribed sequences, BLAST searches uncovered links to protein sequences in only 3 of these 22 microsatellite sequences. Locus Cocl17 matched with a Follistatin-related precursor in Salmo salar, locus Cocl40 matched with a NEDD4-binding like protein and locus Cocl44 matched with an ELAV-like protein 1 in Salmo salar.
Furthermore, from the 31 previously published whitefish microsatellite loci21 and the 5 loci isolated from closely related salmonid species22,23,24,25, we were able to respectively incorporate 11 and 4 loci which displayed polymorphism in 48 normal fish from all lakes in our analysis (Supplementary Table 1). Two loci, CoclLAV32 and Sfo-8-1, developed in whitefish and brook char, respectively, are duplicated and the PCR amplifies 2 unlinked loci each. The number of alleles per locus for this set of loci ranged from 4 to 21 ( , σ = 5.92). Expected and observed heterozygosities ranged from 0.13 to 0.78 (
, σ = 5.92). Expected and observed heterozygosities ranged from 0.13 to 0.78 ( , σ = 0.19) and from 0.13 to 0.71 (
, σ = 0.19) and from 0.13 to 0.71 ( , σ = 0.18), respectively. There was no evidence of large allele dropout or genotyping errors due to stutter peaks for 17 loci and no loci deviated from Hardy– Weinberg equilibrium, P > 0.05; Supplementary Table 1). There was no evidence of linkage disequilibrium among any of these loci (P > 0.05). Genotyping was consistent across the 48 samples, which were tested twice and yielded the same genotypes. The multiplex optimisation yielded a total of 8 panels for the 37 primer pairs which, due to the apparent duplication of 3 loci, resulted in the successful amplification of 40 loci. In these three loci we were able to score alleles of the expected size for the original sequence as well as a set of alleles in a different size range (Supplementary Table 1). We were able to design one 3-plex, one 4-plex, three 5-plex and three 6-plex multiplex reactions. Supplementary Table 1 summarises the above results and also provides, for each locus, the number of alleles, the size of the flanking sequences, the number of repeated units in the smallest and largest alleles scored, the polymorphic information content (PIC) the estimated null-allele frequency (NAF) and the GenBank accession numbers.
, σ = 0.18), respectively. There was no evidence of large allele dropout or genotyping errors due to stutter peaks for 17 loci and no loci deviated from Hardy– Weinberg equilibrium, P > 0.05; Supplementary Table 1). There was no evidence of linkage disequilibrium among any of these loci (P > 0.05). Genotyping was consistent across the 48 samples, which were tested twice and yielded the same genotypes. The multiplex optimisation yielded a total of 8 panels for the 37 primer pairs which, due to the apparent duplication of 3 loci, resulted in the successful amplification of 40 loci. In these three loci we were able to score alleles of the expected size for the original sequence as well as a set of alleles in a different size range (Supplementary Table 1). We were able to design one 3-plex, one 4-plex, three 5-plex and three 6-plex multiplex reactions. Supplementary Table 1 summarises the above results and also provides, for each locus, the number of alleles, the size of the flanking sequences, the number of repeated units in the smallest and largest alleles scored, the polymorphic information content (PIC) the estimated null-allele frequency (NAF) and the GenBank accession numbers.
Genotype scoring and population analyses
We then validated the suitability of the loci included in the multiplex panels for population genetics analyses by genotyping a total of 494 normal and dwarf whitefish individuals from 5 lakes. Locus Cocl8 did not amplify consistently with samples from Cliff Lake and was excluded from this analysis. From individuals genotyped as controls in each plate, we calculated a mistyping rate of 0.0019 per allele genotyped. Type A and type B error rates were 0 and 0.14% for multiplex 1, 0 and 0.82% for multiplex 2, 0.03% and 0.65%for multiplex 3, 0.22% and 0.07% for multiplex 4, 0.16% and 0.96% for multiplex 5, 0.37% and 1.05% for multiplex 6, 0.56% and 0.78% for multiplex 7 and 0 and 0.03% for multiplex 8. The frequencies of cases that could not be scored were slightly higher. They ranged from 0.2% to 2.6% across all loci and were mainly caused by excessive stuttering of some long alleles at loci Cocl14 and Cocl28. These two loci also showed significant heterozygosities deficits in at least 8 populations. For the remaining loci, most populations were in Hardy-Weinberg equilibrium with a few exceptions. Supplementary Table 2 summarises the genetic diversity found in each locus and population. Genotypic linkage disequilibrium was found in only one out of 7410 pairwise comparisons. Loci Cocl11 and Cocl20 were significantly linked in 9 out of 10 population/loci-pair pairwise comparison (P < 0.05 after Bonferroni correction). For the remaining pairwise comparisons, apparent linkage was found in 1.66% of the total (123 population/loci-pair), which is well below the number of significant comparisons expected by chance. Therefore, we can conclude that 38 out of 39 loci are effectively unlinked to each other.
Fixation indices between ecotypes within lakes based on the infinite allele model (θ) were all significantly different from zero (Table 2) but, as expected, the amplitude of differentiation varied among lakes. According to our predictions, lakes East and Témiscouata showed the least ecotype pair divergence (θ = 0.029, 0.021 respectively) together with Webster Lake (θ = 0.025), while Indian Pond and Cliff Lake showed the most ecotype pair divergence (θ = 0.091 and 0.273 respectively). Finally, according with our predictions, the population phenograms, constructed with the Neighbour Joining algorithm using DCE, DA, (δμ)2 and the maximum likelihood phenogram invariably depicted clusters of normal and dwarf ecotypes within lakes for the least divergent pairs with relatively high bootstrap support (>60%) (data not shown). However, when the most divergent ecotypes from Cliff Lake were included in the analysis, we consistently recovered as sister taxa only the ecotypes from lakes Témiscouata and East and to a lesser extent Webster (Figure 3).
Neighbor-joining (A–C) and Maximum-likelihood (D) unrooted population phenograms estimating relationships among dwarf (D) and normal (N) white fish from five lakes, Témiscouata, East, Indian, Webster and Cliff of the Saint John River.
A) Cavalli-Sforza and Edwards' chord distance (DCE), (B) Nei's distance DA (C) Delta mu (δμ)2 and (D) Maximum-likelihood algorithm. Percentages values based on a thousand bootstraps are given for nodes with at least 60% support.
Discussion
Before the advent of next-generation sequencing technologies, no genetic marker has found such widespread use as microsatellites in the last two decades. This study joins an ever increasing number of others that have successfully employed NGS to detect a virtually unlimited number of polymorphic microsatellites across a wide variety of taxa. Our study takes advantage of Roche-454 titanium chemistry, Illumina®, PE high-throughput sequencing technology and online databases to uncover up to 32,442 microsatellite loci in readily available EST of whitefish. Microsatellite-containing EST have also been reported across a broad range of taxa in recent years26,27,28. Together, these studies demonstrate that the main obstacle of costly de novo isolation for using microsatellite markers for population studies and characterisation has been rapidly overcome by the advent of NGS technologies. For instance, a search in the Web of KnowledgeSM made from January 2009 to October 2013 using ‘next-generation-sequencing and microsatellite*' in the title, abstract or key words as the query, generated 265 hits for publications that developed SSR using NGS while, it only generated one hit previous to 2009. This clearly illustrates the importance and widespread use of this new technology in generating molecular markers. More importantly however, is the fact that a growing proportion of research projects working on modest financial resources are now capable of developing a suitable number of microsatellite markers at minimal laboratory costs and for virtually any taxa6. For example, in this study, we used 6,727 cDNA contigs available online20 to identify 445 microsatellite PAL, of which we were able to optimise 25 new loci in multiplex panels. The number of microsatellite loci detected in this study illustrates how abundant these sequences are in eukaryotic genomes, including expressed sequences. Comparing the number of loci detected across studies and taxa is inherently difficult because genomes vary substantially in their frequency of microsatellites and genome size6. However, the number of loci detected here favourably compares with other studies e.g.26,27,29. Irrespective of the difficulty to compare microsatellite abundance in different taxa, an important message of our study is that a virtually unlimited number of microsatellite loci can be identified as by-product of transcriptome characterization projects, which are ever more available.
The relative yield of microsatellite-containing EST from the two NGS platforms was very different. In the early years of the application of NGS technologies for marker development, it was pointed out that the Roche-454 sequencing technology was probably more suited for non-model species since no reference genome was available and the technology produced longer fragments, leading to easier contig assembly e.g.30. More recently, using this technology, Schoebel, et al.6 carried an extensive survey of microsatellite abundance in 17 non-model species including plants, fungi, invertebrates, birds and mammals and determined if flanking regions were suitable for primer development. They concluded that depending on the species, a different amount of 454 pyrosequencing data might be required for successful identification of a sufficient number of microsatellite markers for ecological genetic studies. Our study shows that the greater coverage offered by the Illumina® platform, combined with the production of PE sequences, greatly overcomes this difficulty and facilitates contig assembly. In our case, this resulted in up to 4.2 times more contigs and 16.6 times more microsatellite PAL from our Illumina® dataset than from the Roche-454 dataset and this with comparable investment in time and money. Compared to the months that used to be necessary to obtain a dozen of microsatellite loci through more traditional approaches, such as cloning of enriched libraries and Sanger sequencing, the thousands of loci identified here required only 2 days of work once the EST datasets were available and the time required to obtain NGS data for most species using a large scale sequencing platform is considerably shorter than with previous methods. For example, even when large sequence datasets are not available, the construction and sequencing of PE RNA libraries usually takes less than a month and provides not only a virtually unlimited amount of microsatellite markers but also valuable data for studying gene expression patterns involved in a particular physiological condition or a stage of development of any organism.
Most of the markers of our multiplex reactions, optimised using 48 normal fish were highly polymorphic, conformed to Hardy-Weinberg equilibrium and are physically unlinked. However, two loci had large heterozygous deficits, which was probably caused by null alleles or large allele dropout and two other loci are most likely physically linked. To better estimate the frequency of null alleles, we relied on the software cervus 3.031. The method searches for loci with a significant homozygote excess evenly distributed across all homozygote classes. Loci Cocl14 and Cocl28 had null-allele frequencies of 0.75 and 0.27, respectively, suggesting that these loci either have null alleles, large allele dropout or the level of stuttering hampers the reliable scoring of alleles differing within one or two repeated units. For the loci with positive null-allele frequencies (n = 12), our estimates ranged between 0% and 13% ( , σ = 0.04) and only two loci had a null-allele frequency above 10% (Supplementary Table 1). The remaining loci (n = 25) displayed negative null-allele frequencies, did not deviate from HWE and are thus unlikely to be affected by any form of genotyping errors or mismatches in the 3′-end of the primer sequences. Simulation studies suggest that null alleles with frequencies between 5% and 8% should have only minor effects on classical estimates of population differentiation32. Hence, the genotyping results for at least 36 of the developed loci could be considered robust enough for most population genetics and demographic history applications.
, σ = 0.04) and only two loci had a null-allele frequency above 10% (Supplementary Table 1). The remaining loci (n = 25) displayed negative null-allele frequencies, did not deviate from HWE and are thus unlikely to be affected by any form of genotyping errors or mismatches in the 3′-end of the primer sequences. Simulation studies suggest that null alleles with frequencies between 5% and 8% should have only minor effects on classical estimates of population differentiation32. Hence, the genotyping results for at least 36 of the developed loci could be considered robust enough for most population genetics and demographic history applications.
Although we attempted to incorporate 31 loci that were previously developed from size-selected microsatellite-enriched libraries of total genomic DNA into the analysis, we were only able to successfully amplify 11 of those loci in the 48 normal fish samples used during the development phase. Rogers, et al.21 Assessed locus variability using only 10 samples of C. clupeaformis originating from East Lake (n = 3), Cliff Lake (n = 4) and Temiscouata Lake (n = 3). We initially used 9 different samples from each lake for these three lakes and 18 additional samples from Webster Lake and Indian Pond (9 each). It is difficult to ascertain why we only had a 30% rate of success in amplifying and successfully scoring loci that had been previously published and tested. There are however, a couple of possible explanations. First, a simple explanation would be that the addition of the M13 tail in these loci prevents the efficient PCR amplification of the targeted DNA fragment. Testing this possibility is however beyond the scope of this study and the cost of synthesising fluorescently labelled primers is both very high and unnecessary since we have sufficient amplifying loci from the other data bases. Secondly, the genotyping of alleles were performed with different technologies. We used M13 6-FAM fluorescently labelled primers and an ABI 3130x Genetic Analyser, while Rogers, et al.21 used direct incorporation of fluorescently labelled dUTP-TAMRA to score the genotypes on a FMBIO II scanner (Hitachi). It is possible the latter method of genotype visualization in a small number of samples does not permit a full appreciation of the potential genotyping difficulties in multiplex reactions.
Our results confirmed the findings of previous studies that have shown that, although there is reproductive isolation between dwarf and normal whitefish within lakes, there is also a mosaic of genetic differentiation levels between ecotypes among different lakes13,14,15, which can range from 1% to 54% depending on the mutation model used to calculate fixation indices. Fixation indices based on the SMM (RST) were generally higher than IAM (FST) estimates. The IAM model makes the assumption that each mutation gives rise to a unique allelic state with no known relationship to other allelic states. Any similarity between samples is then attributable to a recent divergence from a common ancestor or due to migration. Since there is evidence that most microsatellites generally mutate in a step-wise fashion33, but see also Ellegren2 and therefore do not lose information about ancestral allelic states, the IAM estimates using microsatellite data tend to overestimate genetic similarity and underestimate coalescence times. However, the performance of IAM improves if populations have only recently diverged, as in this case, it is expected that genetic drift rather than new mutations would be the main process creating differentiation. The generally higher values of RST compared to FST observed in this study therefore suggest that the observed population structure is not of very recent origin (circa hundreds to a few thousands generations) and that drift and restricted migration are not the only processes that have led to the genetic differentiation. Mutational events are likely to have also played a role in this differentiation. Our results also confirmed the hypothesis that lakes Témiscouata and East harbour the least divergent ecotype pairs and Cliff harbours the most divergent pair. Irrespective of the mutation model used to estimate the fixation indices, our values are comparable to those obtained by Lu and Bernatchez14 using only 6 loci, Campbell and Bernatchez34 using 440 amplified fragment length polymorphism (AFLP) loci and Renaut, et al.13 based on 94 single nucleotide polymorphic (SNP) nuclear loci for the same group of lakes (Table 2). Their fixation index estimates ranged from 1 to 8.5% for East and Témiscouata and from 22 to 39% for Cliff. Interestingly, a recent study that performed a genome scan using 2734 SNP from mapped loci to obtain a genome-wide average FST (μGW) found extremely close values of the ones we estimated with this 39 loci15.
Finally, we also confirmed the hypothesis that population phenograms should recover sister taxa clusters for the least divergent population pairs but not necessarily for the most divergent pairs. Different levels of admixture within lakes would result in different apparent scenarios of evolutionary relationships within and among lakes. While high levels of admixture between the Acadian and Atlantic postglacial lineages would result in an apparent sister taxa relationship, complete reproductive isolation as observed in Cliff Lake may result in a blurred signal of evolutionary relationships among lakes as a result of size homoplasy at microsatellite loci e.g.35. However, there is clear evidence that secondary contact between Acadian and Atlantic glacial lineages clearly occurred in Cliff, Indian and Webster lakes, but the historical signal of strong differentiation between founding lineages has been maintained only in Cliff Lake. Our results fully concur with those of Gagnaire, et al.15 suggesting that Indian Pond is less admixed than Webster, East and Témiscouata lakes. The microsatellite generated distances obtained with the loci we genotyped, clearly show that irrespective of the method used to depict evolutionary relationships for all populations, invariably the dwarf ecotype of Indian and Cliff lakes cluster as sister taxa showing that they retain the signal of common origin from the same lineage. On the other hand, for East Lake our results further substantiate that a single founding population from the Acadian lineage invaded this newly created habitat. Finally, for Témiscouata Lake, further historical demographic analyses are necessary to reveal its colonisation patterns. In fact, the microsatellites developed here are expected to reveal the demographic history of this adaptive radiation in both the timing and intensity of demographic events (i.e., the extent to which these populations increased or decreased in size and how gene flow varied in time between them). Therefore an important gap in our understanding of the diversification of these fishes is the lack of documentation of the level of parallelism that may exist in the demographic history of this adaptive radiation. A powerful approach to disentangle these complex demographic processes is the use of microsatellite generated distances to draw inferences on historic demography using the observed distribution of genetic variation in contemporary populations36,37
This work represents a major advance in the identification of large numbers of informative SSR loci in whitefish, an increasingly important species in the study of the mechanisms that generate and maintain biodiversity. Using the 454 GS FLX Titanium and Illumina® sequencing technologies, we described the first 22 microsatellite loci found in EST sequences in lake whitefish. Furthermore, we optimised and validated a total of 8 multiplex panels encompassing 39 loci for the study of whitefish evolution and demographic history.
Methods
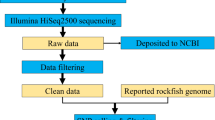
The normal and dwarf whitefish samples used for population analyses in this study have been previously used in two other studies where the sampling and DNA extraction procedures are described38,39. Transcriptome sequences were obtained from the results of two previous studies. First, we used a Roche-454 sequencing transcriptome dataset published in Renaut, et al.19. Second, additional transcriptome sequences were obtained from an unpublished dataset generated to characterise the transcriptome of dwarf and normal whitefish hybrids and backcrosses (AM Dion-Côté and L Bernatchez, unpublished). The dataset consisted of 628,416,550 PE reads. Tags and adaptor sequences were removed and reads were then trimmed, using only reads with 90 or more base pairs with a quality threshold of 2% error-rate per base and a maximum of one N nucleotide. The assembly was performed using the software ABySS40 and the CLC Genomics Workbench 3.1 with similarity set to 0.97 and overlap to 0.5.
Contig assemblies and microsatellite detection
To uncover sequences containing microsatellites in the whitefish transcriptome, we used three datasets: 1) 627,608 reads generated by Roche-454 sequencing and assembled into 14,449 contigs, as reported in19, 2) 6,727 cDNA contigs of the Genomics Research on All Salmon Project (cGRASP) available online20 and 3) 628,416,550 PE reads generated by Illumina® HiSeq™ Sequencing Systems assembled in 75,905 contigs (AM Dion-Côté and L Bernatchez, unpublished). To identify sequences containing microsatellites, we used the mreps software which has been designed to isolate and characterise highly polymorphic microsatellite loci in a comparative genomic or transcriptomic context41. For the purpose of evaluating the number of microsatellite-containing EST from the different datasets, we defined 3 filtering steps (Figure 1). In the first step, we included loci containing di-, tri-, tetra- penta-, sexta-, septa- and octa-nucleotides SSR with 20 to 160 base pairs of repeated units (i.e. 10 tandem repeats for di-, 6 for tri-, 5 for tetra-nucleotides, etc) and a maximum proportion of non-perfect repeats of 0.333. Since several thousands of potential loci were detected in this filtering step and the most commonly used microsatellites in population studies are di- tri- and tetra-nucleotides, we retained only this type of loci in the third filtering step. Furthermore, as microsatellite polymorphism increases with the number of microsatellite repeats42 and in order to increase the chances of finding variable markers, we also focused on microsatellites with at least 10, 8, or 6 tandem repeats of di- tri- and tetra-nucleotides, respectively. Finally, since only loci with sufficiently large flanking sequences around the microsatellite are potentially useful for primer design for successful PCR amplification, we selected loci with a minimum of 40 flanking nucleotides on each side of the tandem repeated units. The scripts used in the filtering steps are available upon request. These loci are referred as “Potentially Amplifiable Loci”, or PAL5. Figure 2 depicts the total number of PAL identified in 97,081 EST contigs of whitefish for the three classes of repeats (di- tri- and tetra-nucleotide microsatellite. To annotate these PAL, we blasted the contigs containing them against the Swissprot database43. From the 11,006 PAL identified, we selected 44 sequences for subsequent analyses. The selection criteria for these loci were simple. Due to budget constraints and because our aim was to obtain 30 to 40 loci, of the thousands of microsatellites containing EST identified in these analyses, we only selected the first 44 loci of the data bases that were suitable for the design of primers, had sufficient number of repeats for subsequent analyses and had sequence lengths from 100 to 400 base pairs to accommodate several loci during multiplexing. As we found sufficient loci among these and the previously published microsatellite, we proceeded with these loci for the subsequent analyses. Primer design was performed using the on-line version of Primer 344. These were synthesised with the universal M13 primer in the 5′ end of the forward primer for their later analysis using a fluorescently labelled M13 primer according to Schuelke45.
Multiplex optimisation
PCR amplifications were performed in a reaction volume of 20 μl, containing 20 ng of genomic (gDNA), 0.5 pmol of each primer, 75 μM of each dNTP, 1.5 mM of MgCl2, 1 × PCR Green Master Mix buffer (Promega, Madison, WI USA) and 0.5 units of Taq polymerase (Promega). The PCR amplifications were done on an Applied Biosystems 9700 DNA thermal cycler using the following conditions: an initial denaturation step at 95°C for 10 min followed by 35 cycles of 30 s at 94°C, 1 min at either 58 or 54°C, depending on the primer's melting point and 1 min 30 s at 72°C and a last cycle of extension at 72°C of 10 min. The amplification products were visualized on agarose. A total of 31 primer pairs amplified a clear PCR product and were selected for assessing their allelic size ranges using 48 normal individuals from all lakes. The rationale for using samples from all lakes is that by genotyping diverse non interbreeding populations, we aimed to detect a wide spectrum of the genetic diversity present in this species complex and thus help in the subsequent design of the multiplex panels. The PCR products were then separated on a capillary sequencer (ABI 3130x Genetic Analyzer, Applied Biosystems, USA) using GeneScan™ 500 LIZ® Size Standard. Allele sizes were determined with the Gene Mapper® V4.0 program (Applied Biosystems, USA). From this analysis, we identified 22 loci that produced clear PCR products. The forward primer of each of these loci was then synthesised again incorporating in the 5' end one of the four fluorescent markers 6FAM™, VIC®, NED™ and PET® for multiplex PCR analysis. In addition of these 22 microsatellites, we screened 31 previously published whitefish loci21 and 5 loci from closely related salmonid species22,23,24,25.
For the multiplex reactions, we used the Qiagen® Multiplex PCR Kit (Qiagen, Canada). Final volumes and concentrations of the master mixes were optimised to reduce the total genotyping cost18. PCR reactions were done in a final volume of 10 μL for all multiplexes, with 3 μL of primer mix, 5 μL of Qiagen Multiplex Mix and 2 μL of gDNA template (10–15 ng/μL). The cycling conditions for the eight multiplexes differed only in the annealing temperature: an initial step at 95°C for 15 min; then 35 cycles at 94°C for 30 s, 54 to 61°C for 3 min and 72°C for 1 min; and a final elongation step at 60°C for 30 min. Table 1 summarises the information about the primers sequences, concentrations in the primer premix [μM], GenBank accession numbers and multiplex panel annealing temperatures.
Genotype scoring and analyses
Once multiplex conditions were optimised, we screened a total of 494 normal and dwarf whitefish from 5 different lakes for variation at 39 polymorphic microsatellite loci. In order to ensure that the allele spread calibration held for each set of samples analysed, we always included the same 6 individuals in each plate to be genotyped as reference standards. For each marker, clear reading rules were defined and illustrated using screen shots of the fluorogramas that were provided to a second scorer to ensure scoring consistency. An error rate was obtained by comparing scoring across scorers at a subset of 200 individuals. Two types of errors were distinguished. Type A corresponds to cases where reader 1 called a genotype as heterozygous and reader 2 typed it as homozygous, or vice versa. Type B corresponds to cases where a wrong allele was called by one of the scorers5. For each marker, the total number of alleles (NA), the observed (HO) and expected (HE) heterozygosities, the polymorphism information content (PIC) and the null-allele frequency (NAF) were determined using the program CERVUS 3.031. Samples were also tested for linkage disequilibrium and departure from Hardy-Weinberg equilibrium using Arlequin V 3.546.
A further test of the scoring's robustness was carried out by establishing the evolutionary relationships between ecotypes within lakes and among all five lakes. As stated above, the recent origin and reproductive isolation with reduced gene flow for these species pairs has already been demonstrated using different methods11,13,14,15,17,47. These studies have unambiguously established that East and Témiscouata lakes harbour the normal and dwarf ecotype pairs that show the least differentiation and that they may have experienced recent introgression (FST < 0.05), while Cliff Lake harbours the most differentiated pair (FST ≈ 0.25)13,14,15,47. It is therefore reasonable to hypothesise that the evolutionary relationships depicted with this microsatellite dataset should support the sister taxa status for the dwarf and normal ecotypes of lakes East and Témiscouata. To test this hypothesis, we first investigated the amplitude of genetic differentiation between ecotypes within lakes. Arlequin V 3.546 was used to calculate weighted FST (θ) statistics by estimating pairwise fixation indices based on allele frequency variation over all loci. Subsequently, we established the evolutionary relationships among ecotypes and lakes using different approaches4. To evaluate the performance of model-specific distance estimators, we followed the procedure described in48. A flowchart summarising the procedures followed to identify and validate the microsatellite loci used in this study is presented in Supplementary Figure 1.
References
Miesfeld, R., Krystal, M. & Arnheim, N. A member of a new repeated sequence family which is conserved throughout eukaryotic evolution is found between the human Delta-globin and Beta-globin genes. Nucleic Acids Res. 9, 5931–5947, 10.1093/nar/9.22.5931 (1981).
Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–445, 10.1038/nrg1348 (2004).
Estoup, A., Beaumont, M., Sennedot, F., Moritz, C. & Cornuet, J. M. Genetic analysis of complex demographic scenarios: Spatially expanding populations of the cane toad, Bufo marinus. Evolution 58, 2021–2036, 10.1554/03-584 (2004).
Takezaki, N. & Nei, M. Empirical tests of the reliability of phylogenetic trees constructed with microsatellite DNA. Genetics 178, 385–392, 10.1534/genetics.107.081505 (2008).
Castoe, T. A. et al. Thousands of microsatellite loci from the venomous coralsnake Micrurus fulvius and variability of select loci across populations and related species. Mol. Ecol. Res. 12, 1105–1113, 10.1111/1755-0998.12000 (2012).
Schoebel, C. N. et al. Lessons learned from microsatellite development for nonmodel organisms using 454 pyrosequencing. J.Evolution. Biol. 26, 1–12, 10.1111/jeb.12077 (2013).
Aguilar-Espinoza, A., Guzman-Riffo, B., Haye, P. A. & Gallardo-Escarate, C. Mining of EST-SSR from 454 pyrosequencing in the surf clam Mesodesma donacium (Lamark, 1818). Conserv. Genet. Res. 4, 829–832, 10.1007/s12686-012-9652-3 (2012).
Bernatchez, L. et al. On the origin of species: insights from the ecological genomics of lake whitefish. Philos. Trans. R. Soc. B-Biol. Sci. 365, 1783–1800, 10.1098/rstb.2009.0274 (2010).
Bernatchez, L. & Dodson, J. J. Allopatric origin of sympatric populations of lake whitefish (Coregonus-clupeaformis) as revealed by mitochondrial-dna restriction analysis. Evolution 44, 1263–1271, 10.2307/2409287 (1990).
Bernatchez, L. & Dodson, J. J. Phylogenic relationships among palearctic and nearctic whitefish (Coregonus spp) populations as revealed by mtDNA variation. Can. J. Fish. Aquat. Sci. 52, 1595–1595, 10.1139/f95-752, (1995).
Pigeon, D., Chouinard, A. & Bernatchez, L. Multiple modes of speciation involved in the parallel evolution of sympatric morphotypes of lake whitefish (Coregonus clupeaformis, Salmonidae). Evolution 51, 196–205, 10.2307/2410973 (1997).
Bernatchez, L., Chouinard, A. & Lu, G. Q. Integrating molecular genetics and ecology in studies of adaptive radiation: whitefish, Coregonus sp., as a case study. Biol. J. Linnean Soc. 68, 173–194, 10.1111/j.1095-8312.1999.tb01165.x (1999).
Renaut, S., Nolte, A. W., Rogers, S. M., Derome, N. & Bernatchez, L. SNP signatures of selection on standing genetic variation and their association with adaptive phenotypes along gradients of ecological speciation in lake whitefish species pairs (Coregonus spp.). Mol. Ecol. 20, 545–559, 10.1111/j.1365-294X.2010.04952.x (2011).
Lu, G. Q. & Bernatchez, L. Correlated trophic specialization and genetic divergence in sympatric lake whitefish ecotypes (Coregonus clupeaformis): Support for the ecological speciation hypothesis. Evolution 53, 1491–1505, 10.2307/2640895 (1999).
Gagnaire, P. A., Pavey, S. A., Normandeau, E. & Bernatchez, L. The genetic architecture of reproductive isolation during speciation-with-gene-flow in lake whitefish species pairs assessed by RAD-sequencing. Evolution 67, 2483–2497, 10.1111/evo.12075 (2013).
Landry, L., Vincent, W. F. & Bernatchez, L. Parallel evolution of lake whitefish dwarf ecotypes in association with limnological features of their adaptive landscape. J.Evolution. Biol. 20, 971–984, 10.1111/j.1420-9101.2007.01304.x (2007).
Landry, L. & Bernatchez, L. Role of epibenthic resource opportunities in the parallel evolution of lake whitefish species pairs (Coregonus sp.). J.Evolution. Biol. 23, 2602–2613, 10.1111/j.1420-9101.2010.02121.x (2010).
Guichoux, E. et al. Current trends in microsatellite genotyping. Mol. Ecol. Res. 11, 591–611, 10.1111/j.1755-0998.2011.03014.x (2011).
Renaut, S., Nolte, A. W. & Bernatchez, L. Mining transcriptome sequences towards identifying adaptive single nucleotide polymorphisms in lake whitefish species pairs (Coregonus spp. Salmonidae). Mol. Ecol. 19, 115–131, 10.1111/j.1365-294X.2009.04477.x (2010).
Consorcium for Genomics Research on All Salmon Project cGRASPhttp://web.uvic.ca/grasp/ (October 2013).
Rogers, S. M., Marchand, M. H. & Bernatchez, L. Isolation, characterization and cross-salmonid amplification of 31 microsatellite loci in the lake whitefish (Coregonus clupeaformis, Mitchill). Mol. Ecol. Notes 4, 89–92, 10.1046/j.1471-8286.2003.00578.x (2004).
Turgeon, J., Estoup, A. & Bernatchez, L. Species flock in the North American Great Lakes: Mol. Ecol. of Lake Nipigon Ciscoes (Teleostei: Coregonidae: Coregonus). Evolution 53, 1857–1871, 10.2307/2640446 (1999).
Patton, J. C., Gallaway, B. J., Fechhelm, R. G. & Cronin, M. A. Genetic variation of microsatellite and mitochondrial DNA markers in broad whitefish (Coregonus nasus) in the Colville and Sagavanirktok rivers in northern Alaska. Can. J. Fish. Aquat. Sci. 54, 1548–1556, 10.1139/cjfas-54-7-1548 (1997).
Saint-Laurent, R., Legault, M. & Bernatchez, L. Divergent selection maintains adaptive differentiation despite high gene flow between sympatric rainbow smelt ecotypes (Osmerus mordax Mitchill). Mol. Ecol. 12, 315–330, 10.1046/j.1365-294X.2003.01735.x (2003).
Angers, B., Bernatchez, L., Angers, A. & Desgroseillers, L. Specific microsatellite loci for brook charr reveal strong population subdivision on a microgeographic scale. J. Fish Biol. 47, 177–185, 10.1111/j.1095-8649.1995.tb06054.x (1995).
Wang, R. et al. Rapid development of molecular resources for a freshwater mussel, Villosa lienosa (Bivalvia: Unionidae), using an RNA-seq-based approach. Freshw. Sci. 31, 695–708, 10.1899/11-149.1 (2012).
Wang, B. et al. Transcriptome sequencing of black grouse (Tetrao tetrix) for immune gene discovery and microsatellite development. Open Biol 2, 10.1098/rsob.120054 (2012).
Santure, A. W., Gratten, J., Mossman, J. A., Sheldon, B. C. & Slate, J. Characterisation of the transcriptome of a wild great tit Parus major population by next generation sequencing. BMC Genomics 12, 10.1186/1471-2164-12-283 (2011).
Zhang, J. et al. De novo assembly and Characterisation of the Transcriptome during seed development and generation of genic-SSR markers in Peanut (Arachis hypogaea L.). BMC Genomics 13, 10.1186/1471-2164-13-90 (2012).
Vera, J. C. et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol. Ecol. 17, 1636–1647, 10.1111/j.1365-294X.2008.03666.x (2008).
Kalinowski, S. T., Taper, M. L. & Marshall, T. C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106, 10.1111/j.1365-294X.2007.03089.x (2007).
Chapuis, M.-P. & Estoup, A. Microsatellite null alleles and estimation of population differentiation. Mol. Biol. Evol. 24, 621–631, 10.1093/molbev/msl191 (2007).
Goldstein, D. B., Linares, A. R., Cavallisforza, L. L. & Feldman, M. W. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc. Natl. Acad. Sci. U. S. A. 92, 6723–6727, 10.1073/pnas.92.15.6723 (1995).
Campbell, D. & Bernatchez, L. Genomic scan using AFLP markers as a means to assess the role of directional selection in the divergence of sympatric whitefish ecotypes. Mol. Biol. Evol. 21, 945–956, 10.1093/molbev/msh101 (2004).
Estoup, A., Jarne, P. & Cornuet, J. M. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol. Ecol. 11, 1591–1604, 10.1046/j.1365-294X.2002.01576.x (2002).
Beaumont, M. A. Detecting population expansion and decline using microsatellites. Genetics 153, 2013–2029 (1999).
Beaumont, M. A. & Rannala, B. The Bayesian revolution in genetics. Nat. Rev. Genet. 5, 251–261 (2004).
Evans, M. L. & Bernatchez, L. Oxidative phosphorylation gene transcription in whitefish species pairs reveals patterns of parallel and nonparallel physiological divergence. J.Evolution. Biol. 25, 1823–1834, 10.1111/j.1420-9101.2012.02570.x (2012).
Evans, M. L., Praebel, K., Peruzzi, S. & Bernatchez, L. Parallelism in the oxygen transport system of the lake whitefish: the role of physiological divergence in ecological speciation. Mol. Ecol. 21, 4038–4050, 10.1111/j.1365-294X.2012.05682.x (2012).
Simpson, J. T. et al. ABySS: A parallel assembler for short read sequence data. Genome Res. 19, 1117–1123, 10.1101/gr.089532.108 (2009).
Kolpakov, R., Bana, G. & Kucherov, G. mreps: efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res. 31, 3672–3678, 10.1093/nar/gkg617 (2003).
Weber, J. L. Informativeness of human (dc-da)n.(dg-dt)n polymorphisms. Genomics 7, 524–530, 10.1016/0888-7543(90)90195-z (1990).
GenBank. NIH Genetic Sequence Database http://www.ncbi.nlm.nih.gov/genbank/ (October 2013).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods in molecular biology (Clifton, N.J.) 132, 365–386 (2000).
Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotech. 18, 233–234, 10.1038/72708 (2000).
Excoffier, L., Laval, G. & Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50 (2005).
Renaut, S. et al. Genome-wide patterns of divergence during speciation: the lake whitefish case study. Philos. Trans. R. Soc. B-Biol. Sci. 367, 354–363, 10.1098/rstb.2011.0197 (2012).
Rico, C. et al. No evidence for parallel sympatric speciation in cichlid species of the genus Pseudotropheus from north-western Lake Malawi. J.Evolution. Biol. 16, 37–46 (2003).
Acknowledgements
C.R. gratefully acknowledges the Spanish Ministry of Education (Grant № PR2010-0601) and the Spanish Scientific Research Council (CSIC) for the financial support received during his stay at L.B. laboratory. C.R. also acknowledges F. Hiraldo Cano and J.J. Negro Balsameda for their support and encouragement during his stay in Canada. We are also grateful to M.L. Evans and W. Adam for sampling whitefish. This research was financially supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant and a Canadian Research Chair to L.B.
Author information
Authors and Affiliations
Contributions
C.R. and L.B. designed the research. C.R. performed primer design, most laboratory tasks, microsatellite genotyping, population genetic analyses and wrote the first draft of the manuscript, E.N. performed the NGS bioinformatic analysis, A.M.D.C. generated the Illumina libraries and sequences, M.I.R. and G.C. assisted in designing experiment, all laboratory task and genotyping of SSR, all authors assisted in preparing the manuscript and read and approved the final version.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Tables S1 and S2 and Figure S1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Rico, C., Normandeau, E., Dion-Côté, AM. et al. Combining next-generation sequencing and online databases for microsatellite development in non-model organisms. Sci Rep 3, 3376 (2013). https://doi.org/10.1038/srep03376
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03376
This article is cited by
-
Identification and validation of simple sequence repeats markers in Tilletia indica and compatibility assay of monosporidial lines
Indian Phytopathology (2022)
-
De novo assembly, transcriptome characterization and marker discovery in Indian major carp, Labeo rohita through pyrosequencing
Genetica (2022)
-
Characterization of microsatellites in the endangered snow leopard based on the chromosome-level genome
Mammal Research (2021)
-
Comprehensive genus-wide screening of seahorse microsatellite loci identifies priority species for conservation assessment
Conservation Genetics Resources (2021)
-
Microsatellite development from genome skimming and transcriptome sequencing: comparison of strategies and lessons from frog species
BMC Genomics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.