Abstract
The advent of state-of-the-art brain imaging technologies in recent years and the ability of such technologies to provide high-resolution information at both structural and functional levels has spawned large efforts to introduce novel non-invasive imaging biomarkers for early prediction and diagnosis of brain disorders; however, their utility in both clinic and drug development at their best resolution remains limited to visualizing and monitoring disease progression. Given the fact that efficient translation of valuable information embedded in brain scans into clinical application is of paramount scientific and public health importance, a strategy is needed to bridge the current gap between imaging and molecular biology, particularly in neurodegenerative diseases. As an attempt to address this issue, we present a novel computational method to link readouts of imaging biomarkers to their underlying molecular pathways with the aim of guiding clinical diagnosis, prognosis and even target identification in drug discovery for Alzheimer's disease.
Similar content being viewed by others
Introduction
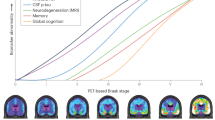
Recent advancements in structural and functional neuroimaging techniques offer unprecedented opportunities to visualize the brain structure and function, to non-invasively monitor the progression of a disease over time, or to track disease trajectories. Different types of imaging reveal different aspects of the brain complexity: Magnetic Resonance Imaging (MRI), Computerized Tomography (CT) and Diffusion Tensor Imaging (DTI) are designed to localize anatomical areas and structures (structural imaging techniques) whereas functional Magnetic Resonance Imaging (fMRI) and Positron Emission Tomography (PET) are used to capture neural activity at molecular level (functional imaging techniques)1,2. Such imaging technologies have been used to identify structural and functional changes associated with different stages of progressive neurodegenerative diseases such as Alzheimer's disease (AD)3.
In AD patients, progressive loss of memory and cognitive abilities is attributed to the dysfunction and death of nerve cells in specific regions of the brain4. Imaging techniques have established the existence of such link between brain structural and functional changes by showing the spatio-temporal patterns of cell death across affected brain regions. Differential patterns of brain atrophy observed in the brain of AD patients with the help of imaging techniques indicate that distribution of particular structural changes in specific regions of the AD brain may reflect the underlying pathology5.
Based on the above-mentioned capabilities of imaging technologies, increasing number of imaging studies has been published on diagnosis and prognosis of AD but the reported applications are still limited to clinical monitoring of anatomical lesions or injuries of brain in the course of the disease. On the other hand, still assessment of persons with brain disorders and diagnostic decision-making process for such patients is highly subjective to the skills of examiner and the patient's abilities, which shows the current limitations of brain imaging techniques for informing the diagnosis beyond the behavioral assessments6. Current automated methods for diagnosis of neuropsychiatric disorders make use of classification algorithms to classify the brain scans of participants based on measurements of local variation in the morphological features of the brain7. Accordingly, the diagnostic information derived from such imaging-based methods is often unspecific and the knowledge behind the molecular mechanism underlying the measured imaging outcomes remains implicit or unknown. In fact, the challenge of linking clinical outcomes to their underlying molecular events has been long of interest to the scientific community and to the pharmaceutical industry as well. This is because it will help to obtain better understanding of the disease mechanism at molecular level, particularly for personalized medicine applications. The technique of molecular imaging using reporter molecules that provide information on particular molecular or cellular events has been around for some while but it is not in clinical use yet and its diagnostic as well as prognostic application will be limited to tracing of single cell or single cellular process8.
To the best of our knowledge, there is no suggestion in the scientific or patent literature preceding the present study how to facilitate diagnosis and prognosis of brain diseases by translating information from a plurality of brain scan images to underlying region-specific disease pathways. Therefore, it would be desirable to provide a method that is able to not only diagnose brain images more accurately with higher specificity to the disease but improve prognosis by prescribing efficient and personalized therapies based on medical history of individual patients. Such a method could be potentially upgraded to a clinical decision-support system that would adjoin neuroimaging softwares. It could also support target identification and biomarker discovery efforts as well.
The present work proposes a novel strategy using an integrative computational approach, which incorporates the information of imaging and potential protein biomarkers specific to disease into a brain-specific protein interaction network. Enrichment analysis for known pathways further validated the method and unveiled the high impact of immune system on the pathology of AD.
Results
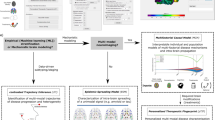
The core methodology for translation of imaging readouts to molecular pathway maps consists of two steps:
Step 1 intends to integrate the information of both imaging and molecular biomarkers into a brain-specific network model (so-called brain interactome), which represents experimentally confirmed protein interactions (i.e. network edges) in 15 anatomical regions of the human brain9. Since reports on brain imaging contain meta-information about clinical specifications of patient subjects such as stage of the disease and the affected region of the brain, the idea is to ultimately generate specific disease subnetworks out of the brain interactome that represent protein interactions in affected regions of the diseased brain. This is achieved by mapping information of imaging outcomes onto the region-specific brain interactome (Figure 1). For validation purposes, we have used imaging information reported in the literature on AD but the source of image-based diagnosis could in essence be extended to the clinician's medical report or any other diagnostic annotation attached to images.
Step 2 involves extraction of identified subnetworks from the brain interactome based on the affected regions that are diagnosed by imaging and consequently their analysis for underlying pathways. The pathway analysis is performed on both the entire subnetworks and potential biomarkers mapped onto these subnetworks separately. Afterwards, pathways that are derived from subnetworks and pathways that are derived from mapped biomarkers are being matched so that potential molecular biomarkers act as “pins” on the disease map to guide the analysis to the core biological processes deemed to drive the pathology of the disease (Figure 2).
Method validation using imaging readouts of Alzheimer's patients
Our semantic information retrieval system, SCAIView, retrieved 5698 PubMed abstracts reporting clinical application of imaging techniques for diagnosis of AD (MRI: 3458, PET: 1989, DTI: 251) and containing information on both AD and affected brain regions (see Methods). After manual inspection of these abstracts and information extraction, it turned out that the reported brain regions injured in AD could be assigned to three stages of the disease, namely early AD/mild cognitive impairment (MCI), moderate Alzheimer's and advanced Alzheimer's. It was evident from the frequency of AD imaging publications that majority of these studies reported the application of imaging methods to diagnosis of the early stage Alzheimer's disease, reflecting the high priority of finding early diagnostics for AD. Moreover, these efforts were heavily biased toward application of MRI techniques amongst others (Supplementary Table 1).
Analysis of the diagnosed anatomical regions in AD brains (Supplementary Table 1) indicated that both structural and functional techniques report limbic system and its anatomical components (i.e. temporal lobe, hippocampus, cingulate, thalamus and corpus callosum) as the affected regions across disease stages. Based on these readouts, a disease progression trend is observed so that it appears with progression of the disease from early to advanced stage, anatomical lesions extend from temporal lobe, entorhinal and precuneus cortices to prefrontal and cerebral cortices. These readouts suggest that temporal lobe lesion and dysfunction is persistent across disease stages. Therefore, we generated a temporal lobe subnetwork model out of the brain interactome and validated our proposed method on this model. The temporal lobe model is represented by a protein-protein interaction (PPI) subnetwork with 2323 nodes and 3587 edges (Supplementary File 1).
Biomarker-pathway coupling for targeted enrichment analysis
In order to spot pathways causally involved in the progression of AD in the temporal lobe subnetwork model, we searched for molecular indicators (potential biomarker candidates) of AD in the literature, extracted a list of such hypothetical AD biomarkers (see Methods) and mapped them onto the temporal lobe subnetwork. Biomarkers are molecular alterations that can be measured in human tissue, cells or fluids and represent direct steps in the causal pathways of a disease10. As a result, 144 potential biomarkers, including inflammatory and non-inflammatory biomarkers, mapped on the temporal lobe subnetwork. Since these potential biomarkers indicate measurable molecular activities under the AD condition, this strategy helps us guide our analysis to those pathways that are more likely to be involved in the disease mechanism at the molecular level. Moreover, it overcomes the challenge of dealing with the large number of significant pathways that are often resulted from pathway enrichment analysis algorithms, particularly when a large number of proteins participating in network models are submitted for analysis.
To this end, we performed separate pathway enrichment analyses on both the subnetwork proteins and the list of mapped biomarkers. Since the enrichment results from mapped biomarkers are used to guide the analysis on the subnetwork model, enrichment analysis on the biomarker list was performed using BioCarta, KEGG and Reactome separately. The same analyses were performed on the subnetwork proteins. In this way, the same set of pathway annotations from each pathway database is used for pathway comparison and the sensitivity of the pathway matching process between biomarker-derived pathways and subnetwork-derived pathways is maximized. The purpose is to find out which pathways are suggested by potential biomarkers to be perturbed in the subnetwork model and which pathways in the subnetwork model correctly represent the disease mechanism.
Such a biomarker-guided pathway analysis showed that imaging biomarkers point to involvement of six pathways in progression of AD, namely HIV-NEF pathway, FAS signaling pathway, IL2RB pathway, keratinocyte pathway, MAPK signaling pathway and immune system signaling (Table I). Figure 3 illustrates HIV-NEF pathway spotted on the temporal lobe subnetwork model. In this model, there are two approved CNS drugs that target two proteins within the HIV-NEF pathway: Triflusal, which targets NFKB1 and is used for treatment of cerebral infarction and prevention of stroke; and Rasagiline, which targets BCL2 and is applied for treatment of idiopathic Parkinson's disease. Such a drug-target-disease pathway landscape informs which therapies already target a disease pathway in a particular brain region. The specificity of the translated model increased even more when the expression information of mapped potential biomarkers (i.e. overexpressed or underexpressed) under AD conditions was also extracted from the literature and incorporated into the subnetwork model (Figure 3 and Supplementary Table 2).
Representation of recovered HIV-NEF pathway and its first neighbor proteins in the temporal lobe network model.
The model has been enhanced with drug-target and biomarker expression information. Circular nodes in yellow show membership to the HIV-NEF pathway; square nodes in pink are approved drugs targeting the recovered pathway; triangle nodes represent potential biomarkers color coded for their expression levels in AD brain (red: over-expression; green: under-expression).
Extended algorithm for translation of personalized imaging information
The presented methodology, if further optimized, is capable of automating computer-aided translation of imaging information for personalized diagnosis and prognosis. We show this capability by extending the proposed methodology to an algorithm, which may be used in conjunction with neuroimaging softwares and aid clinicians in accurate diagnosis and prognosis of patients that undergo imaging assessment (Figure 4).
Information on abnormal or damaged regions of the patient's brain are extracted from the output of the imaging device by clinician and can be given as input to the imaging input unit, which in turn, sends a signal to the translation processing unit for generation of a disease map as a molecular interaction network. This network will be enriched with patient's medical history data and can be visualized through the visualization unit and a graphical user interface containing information about detailed mechanistic diagnosis and prognosis of the disease in a personalized manner.
Discussion
In spite of invaluable contribution of neuroimaging to the understanding of disease progression, its outcome can not be directly used in the context of molecular systems analyses for translational purposes. The presented methodology is a novel approach to integrating brain imaging readouts into a network model of brain molecular interactions, which was validated using the accumulated knowledge on diagnostic neuroimaging of Alzheimer's disease in the literature. In this way, the pictorial information of brain scans that are not amenable to molecular analysis can be incorporated in a region-specific brain interaction network to analyze the resultant mechanistic models and to validate those models in the context of molecular pathways. Consequently, application of this approach to identifying drug targets can have important implications in CNS drug discovery by reducing the risk of drug failure in clinical trials, given the fact that it uses human imaging data instead of poorly disease-mimicking animal data. Even at the level of sample data, a clear advantage of imaging biomarkers over gene expression signatures in neurodegenerative disease research is that they refer to in-vivo observations of regions and tissues in the diseased brain that are directly involved in the disease initiation and progression whereas gene expression signatures only provide a snapshot of perturbed genes, suffer from heterogeneity of cell types and are limited to post-mortem sampling.
Pathway enrichment analysis on the image-translated molecular map of temporal lobe revealed several pathways that were not previously appreciated to be causally involved in the pathogenesis of AD. The advantage of using pathway enrichment analysis is that the collective effect of reported molecular biomarkers under the disease condition is taken into account in the context of disease pathways. When taken together, these pathways unveiled an important aspect of the Alzheimer's pathology: immune system-driven apoptosis. Significant enrichment of temporal lobe subnetwork model and its mapped biomarkers for HIV infection pathway might indicate that signaling pathways to neuronal damage and apoptosis are elicited from the very early stage of AD, which persist over the period of advanced phase. The role of IL2RB and FAS signaling pathway in immune system-mediated apoptosis complements accumulated evidence that MAPK signaling pathways contribute to the pathogenesis of AD through regulation of neural apoptosis11,12,13.
Enhancing these translated models with drug-target and biomarker information – as shown for HIV-NEF pathway model in Figure 3 – can provide added value to physicians and researchers in several ways:
-
it may support more accurate diagnosis based on molecular etiology of the disease, particularly when measured biomarkers from patient are available and can be mapped onto the model to spot disease pathways. This has implications for mechanistic diagnosis of diseases rather than conventional diagnosis solely based on often overlapping symptoms and signs.
-
it may improve prognostic tasks using the drug-target information that is associated to disease pathways. Patient's therapeutic history can complement the model and support prognostic decision-making through incorporation of individual risk factors such as susceptibility background (e.g. APOE genotype) or environmental risk factors (e.g. aging).
-
it may guide target identification through prediction of drug mode of action in the context of affected tissue, disease stage and perturbed pathway. Information of approved drugs and their targets in disease pathways that are already incorporated in the model can support the concept of polypharmacology for discovery and development of next-generation multi-targeting drugs.
-
it may be used for prediction of companion biomarkers that are mechanistically linked to disease etiology, on one hand and to mode of action of approved or experimental drugs, on the other hand.
In summary, the novel integrative methodology presented here provides insight into the underlying molecular mechanisms of disease progression by linking the clinical readouts of imaging techniques to their corresponding molecular events, but this approach has limitations. The inherent issue of network biology is that completeness of molecular network maps is limited to data availability and validity. Therefore, the specificity and sensitivity of the translation process is a function of the completeness of the brain interactome. Another limiting factor is the low resolution of the protein-protein interaction maps in terms of representing other molecular species than proteins and also directionality of the interactions. However, these shortcomings can be overcome through replacement of PPI networks with causal computational models based on BEL (Biological Expression Language; www.openbel.org). BEL-based mechanistic models not only represent all molecular species such as ions or metabolites but also preserve the directionality of interactions.
This method is generic and can be applied to modeling other brain disorders. The authors foresee that the extended algorithm of this methodology, when optimized and fully automated, has the potential to be used as a clinical decision-support tool for personalized diagnosis and prognosis of patients with brain disorders.
Methods
Information retrieval and extraction
With the help of state-of-the-art text mining and knowledge discovery tools, ProMiner14 and SCAIView15 developed at Fraunhofer Institute SCAI, PubMed abstracts were searched using a dedicated biomarker terminology16. The queries were formulated and executed over entire PubMed abstracts on 14.05.2013 in SCAIView search engine (accessible through http://bishop.scai.fraunhofer.de/scaiview/). An example of query formulation is shown below:
(([MeSH Disease:"Alzheimer Disease"]) AND [BioMarker Terminology:"Diffusion tensor imaging"]).
Similarly, with the help of SCAIView and the biomarker terminology, the following query was performed to obtain a list of potential AD biomarkers:
(([MeSH Disease:"Alzheimer Disease"]) AND [BioMarker Terminology Node:"Evidence Marker"]).
The long list of retrieved potential AD biomarkers was filtered for expression evidence and was subjected to manual inspection of sentences.
Imaging abstracts were manually screened by one of the authors (EY) so that context (relevance to both AD and imaging biomarker) as well as content (information detailing the type of imaging biomarker and the affected brain region) of retrieved documents were checked and the relevant information was extracted.
Reconstruction of the temporal lobe subnetwork
A brain-specific protein-protein interaction (PPI) network representing 15 brain regions was reconstructed using the global human protein interaction network9. These regions include amygdala, cerebellum peduncles, cingulate cortex, hypothalamus, medulla oblongata, occipital lobe, parietal lobe, pons, prefrontal cortex, subthalamic nucleus, temporal lobe, thalamus, caudate nucleus, cerebellum and globus pallidus. The PPI network was then filtered for the affected brain regions to obtain region-specific subnetwork for temporal lobe, based on the edge attributes using Cytoscape software17.
Pathway enrichment analysis
Since the affected region in the early stage was the same in the advanced stage, we only analyzed one subnetwork representing temporal lobe in this study. The subnetwork and corresponding mapped molecular biomarkers were subjected to pathway enrichment analysis (GSEA) in MsigDB18. In order to normalize the pathway matching process, GSEA was performed on both potential biomarkers and subnetwork proteins using separate pathway annotation sets, namely BioCarta (www.biocarta.com), KEGG (www.genome.jp/kegg) and Reactome (www.reactome.org). In order to make the more specific enrichment results from the small number of potential biomarkers comparable to the less specific but high dimensional enrichment results from the large number of subnetwork proteins, we considered the top 20 enrichment results for subnetwork proteins so that the sensitivity of the pathway matching process is preserved.
References
Kherlopian, A. R. et al. A review of imaging techniques for systems biology. BMC Syst. Biol. 2, 74 (2008).
Monchi, O., Benali, H., Doyon, J. & Strafella, A. P. Recent advances in neuroimaging methods. Int. J. Biomed. Imaging 2008, 1–2 (2008).
Johnson, K. A., Fox, N. C., Sperling, R. A. & Klunk, W. E. Brain imaging in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2, a006213 (2012).
Mattson, M. P. Pathways towards and away from Alzheimer's disease. Nature 430, 631–639 (2004).
Ashburner, J. et al. Computer-assisted imaging to assess brain structure in healthy and diseased brains. Lancet Neurol. 2, 79–88 (2003).
Coleman, M. R. et al. Towards the routine use of brain imaging to aid the clinical diagnosis of disorders of consciousness. Brain 132, 2541–2552 (2009).
Bansal, R. et al. Anatomical brain images alone can accurately diagnose chronic neuropsychiatric illnesses. PLoS ONE 7, e50698 (2012).
de Backer, M. E. et al. MR-based molecular imaging of the brain: the next frontier. Am. J. Neuroradiol. 31, 1577–1583 (2010).
Bossi, A. & Lehner, B. Tissue specificity and the human protein interaction network. Mol. Sys. Biol. 5, 260 (2009).
Mayeux, R. Biomarkers: potential uses and limitations. NeuroRx. 1, 182–188 (2004).
Baskys, A. & Blaabjerg, M. Understanding regulation of nerve cell death by mGluRs as a method for development of successful neuroprotective strategies. J. Neurol. Sci. 229, 201–209 (2005).
Strasser, A., Jost, P. J. & Nagata, S. The many roles of FAS receptor signaling in the immune system. Immunity 30, 180–192 (2009).
Kim, E. K. & Choi, E. J. Pathological roles of MAPK signaling pathways in human diseases. Bioch. Biophys. Acta 1802, 396–405 (2010).
Hanisch, D., Fundel, K., Mevissen, H. T., Zimmer, R. & Fluck, J. ProMiner: Rule based protein and gene entity recognition. BMC Bioinformatics 6, S14 (2005).
Ivchenko, O. et al. PLIO: an ontology for formal description of protein-ligand interactions. Bioinformatics 27, 1684–1690 (2011).
Younesi, E. et al. Mining biomarker information in biomedical literature. BMC Med. Inform. Decis. 12, 148 (2012).
Killcoyne, S., Carter, G. W., Smith, J. & Boyle, J. Cytoscape: a community-based framework for network modeling. Methods Mol. Biol. 563, 219–239 (2009).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Acknowledgements
We whish to thank Michael Griebel, Carl Vogt, Stephan Springstubbe and staff of Fraunhofer Gesellschaft/Patente und Lizenzen for their administrative and logistic supports.
Author information
Authors and Affiliations
Contributions
E.Y. conceived the overall methodology, designed the experiment, performed the experiment, generated all figures and wrote the paper. M.H.A. provided access to text-mining and knowledge discovery tools and assisted in paper preparation.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Tables 1 & 2
Supplementary Information
Supplementary File 1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Younesi, E., Hofmann-Apitius, M. Biomarker-guided translation of brain imaging into disease pathway models. Sci Rep 3, 3375 (2013). https://doi.org/10.1038/srep03375
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03375
This article is cited by
-
NeuroRDF: semantic integration of highly curated data to prioritize biomarker candidates in Alzheimer's disease
Journal of Biomedical Semantics (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.