Abstract
Increased autoantibody reactivity in plasma from Myelodysplastic Syndromes (MDS) patients may provide novel disease signatures and possible early detection. In a two-stage study we investigated Immunoglobulin G reactivity in plasma from MDS, Acute Myeloid Leukemia post MDS patients and a healthy cohort. In exploratory Stage I we utilized high-throughput protein arrays to identify 35 high-interest proteins showing increased reactivity in patient subgroups compared to healthy controls. In validation Stage II we designed new arrays focusing on 25 of the proteins identified in Stage I and expanded the initial cohort. We validated increased antibody reactivity against AKT3, FCGR3A and ARL8B in patients, which enabled sample classification into stable MDS and healthy individuals. We also detected elevated AKT3 protein levels in MDS patient plasma. The discovery of increased specific autoantibody reactivity in MDS patients, provides molecular signatures for classification, supplementing existing risk categorizations and may enhance diagnostic and prognostic capabilities for MDS.
Similar content being viewed by others
Introduction
Myelodysplastic syndromes (MDS) encompass a diverse range of hematological disorders, with variable clinical outcomes resulting from individual patients' clinical and biological features1,2. MDS pathogenesis involves multifaceted factors, related to intrinsic hematopoietic precursor cell abnormalities. The prevalent shared pathogenesis causing the ineffective hematopoiesis in MDS involves varying degrees of apoptosis of the hematopoietic cell linage3,4,5. Recent genomic approaches have concentrated on the effects of specific gene mutations and their associated signaling pathways and their role in MDS development and outcome, including the tendency of transitioning to more aggressive disease stages6,7. Currently, the prognosis of patient outcomes is greatly facilitated by the establishment of the International Prognostic Scoring System (IPSS8, recently revised as IPSS-R9). The IPSS takes into account multiple clinical markers to classify lower risk patients (Low, Intermediate 1) as having improved prognoses compared to those with higher risk features (Intermediate 2 and High).
Autoantibody reactivity profiles in human plasma have been employed in multiple other disorders, including immune response in severe acute respiratory syndrome10, diabetes11,12, as well as cancer13,14 using protein microarrays. In MDS patients immunologic abnormalities have been observed15. Furthermore, a higher rate of immune related cell abnormalities has been reported in MDS, predominantly in earlier-stage compared to later-stage MDS patients, including altered immune cell subpopulations, namely regulatory16,17 and inhibitory18 T cells. Additionally, disease progression has been found to be concordant with dynamic shortening of telomeres observed in MDS precursors19,20. Short telomeres and DNA damage in hematopoietic precursors, including those from MDS patients, have been associated with cellular protein secretion21.
To further assess disease related abnormalities in autoantibody reactivity and the possibility of an immune related response in MDS patients of various stages, we have utilized high throughput protein arrays that allow the simultaneous monitoring of changes in autoantibody reactivity to thousands of human proteins. Reactive antibody profiling with protein microarray is in principle the same as Enzyme-linked Immunosorbent Assays (ELISA) with the same antigen-primary antibody-secondary antibody format, with additional advantages including 1) a higher throughput and 2) using fluorescent signals from secondary antibodies instead of the less reproducible enzyme-linked chromogenic signals. Protein microarrays have been reported to have higher throughput, sensitivity and a wider detection range compared to traditional ELISA methods in various applications10,22. Our main hypothesis is that MDS elicits specific autoantibody responses and hence we searched for autoantigen biomarkers related to various MDS patient subgroups compared to control plasmas using protein microarray technology (ProtoArrays v. 5 by Invitrogen). We focused on a retrospective classification of subjects into stable MDS patients (s-MDS), which had not transformed into acute myeloid leukemia (AML) for at least 14 months and generally for multiple years, transforming MDS (t-MDS), where patients eventually acquired AML within a 14-month period and AML post MDS (L) where the patients had already transformed to AML, after previously having being classified as MDS patients23. The MDS and AML patients were compared to a healthy cohort of individuals.
Results
The study was conducted in two sequential separate stages: (I) The exploratory stage, in which multiple patient samples and proteins were tested for Immunoglobulin G (IgG) reactivity and (II) the validation stage using a smaller, high-interest subset of the proteins identified in Stage I based on the retrospective classification and expanded to a larger cohort. The use of this focused subset allowed us to utilize the proteins displaying the greatest degree of differential IgG reactivity between patient groups and healthy controls. The different experimental designs are illustrated in Fig. 1a and described in detail with the results further below.
Study Design and Exploratory Stage I Results.
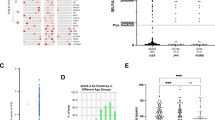
The investigation was carried out in two stages, (a), where in Stage I ProtoArrays were used to identify a high priority set of 35 candidate biomarkers, 25 of which were successfully spotted onto customized arrays for the Stage II focused validation part of the investigation. Stage II identified 3 biomarker candidates, AKT3, ARL8B and FCGR3A, which were also detected using ELISA assays. The 35 candidate biomarkers from Stage I showed distinct higher reactivity in MDS patients compared to the healthy cohort, (b), with higher standardized intensities indicated in yellow, low in blue/turquoise and validated proteins from Stage II marked in red. (c) The binary statistical comparisons between patient subgroups and healthy cohort resulted in different sets of autoantibodies specific to their respective classification. The Venn diagram indicates the overlap between these comparisons, with more candidate biomarkers specific to s-MDS patients than in other subgroups. (d) Various associations, including cancer were discovered using Ingenuity Pathways and Network Analysis (Ingenuity® Systems, http://www.ingenuity.com).
In Stage I multiple plasma samples (75) were obtained from male patients, in the 44–87 (median 70) age range and a healthy cohort (34), in the 52–70 (median 61) age range. As discussed in the Methods, the samples used in our study were obtained early in the patients' courses, to enable the assessment of predictive potential for prolonged clinical courses of the MDS patients (i.e., s-MDS). At the time of sample collection, the patients were classified using the prospective clinical risk-based IPSS system. Following long-term monitoring of the patients, the same samples were also assigned a retrospective classification (into s-MDS, t-MDS, L, as stated above and previously defined23). The patients were compared to a healthy cohort (Table 1a).
After identifying a high-priority set of 35 markers (Fig. 1b–d) in Stage I described below, in the validation Stage II (Fig. 2a) the initial subject pool was enlarged to include both male and female individuals, with 204 patients (119 s-MDS, 42 t-MDS, 43 L) and 112 healthy controls (Table 1b). We note here that differences in median ages between patient and healthy groups were taken into consideration in our Analysis of Variance (ANOVA) model below to ensure they were statistically not a factor for our final results. While Stage I samples were only obtained from male patients, as prior studies in patients with ovarian and prostate carcinoma showed gender differences in antibody reactivity13,14, in Stage II we assessed samples from both genders and found no such differences in our results for MDS patients.
Stage II Design, Analysis and Customized Arrays.
In Stage II focused arrays were analyzed (a), including multiple normalization steps and analysis of variance (ANOVA) to ascertain statistical significance in obtaining a high-priority candidate set of three proteins. Each custom array contained 12 blocks, such as the example (b) and was scanned for Immunoglobulin G, IgG, reactivity [using wavelength 532 nm (green) Laser Emission Fluorescence] and protein levels [GST fusion tag expression levels using wavelength 635 nm (red) Laser Emission Fluorescence], with the superposition of both green and red emission spectra from scanning as shown. The protein arrays from patient signals (c) show increased reactivity to IgG as compared to a negative control. Black and low luminosity indicate no and lower reactivity respectively.
Stage I results
The initial screening of protein arrays containing 9,483 proteins identified 35 autoantigens of high interest showing aberrantly high reactivity in patient subgroups compared to the healthy cohort (Fig. 1b). The Venn diagram (Fig. 1c) indicates minimal overlap between the different retrospective MDS classifications, with the s-MDS group showing the greatest differences compared to the healthy group, which should be noted has greater numbers of available samples. Furthermore, considered as a whole the results support the hypothesis that MDS patients display distinct autoantibody responses compared to a healthy cohort (see also Supplementary Table S1).
This initial screening identified relevant molecular associations, (Ingenuity® IPA Knowledge Database, http://www.ingenuity.com, Fig. 1d) including cancer (14 proteins), apoptosis (12), viral infection (8) and cell movement (8) (Supplementary Table S2). Additionally, 3 proteins are members of the Role of NFAT (nuclear factor of activated T cells) in Regulation of the Immune Response canonical pathway (namely AKT3, FCGR3A and GNAZ) and two are members of the Thyroid Receptor-Retinoid X Receptor (RT/RXR) activation pathway (AKT3, TRH) (Supplementary Table S3, Supplementary Fig. S1).
Stage II results
The high-interest proteins from Stage I (see Methods for details) were tested for validation, using custom protein arrays, with 25 proteins successfully produced and spotted (Fig. 2). To take into account multiple sources of variability, we employed an ANOVA model and filtered results to eliminate effects due to printing, replication and batch effect and subject age (see Methods; Fig. 3a). We identified 3 proteins, AKT3, FCGR3A and ARL8B (Fig. 3b), showing aberrantly increased reactivity in patients compared to healthy (P < 0.01, Bonferroni corrected) and with no other factors considered having a significant confounding effect. The corresponding binary group comparisons of mean differences were also statistically significant [Mann-Whitney U-tests, with 1) AKT3 mean intensities P < 3 × 10−6, <3 × 10−3, <4 × 10−3 for s-MDS, t-MDS and L versus healthy respectively, 2) FCGR3A mean intensities P < 8 × 10−4, <7 × 10−3, for s-MDS and L versus healthy respectively and 3) ARL8B mean intensities P < 4 × 10−5, <2 × 10−3, <3 × 10−6, for s-MDS, t-MDS and L versus healthy respectively].
Stage II Analysis of Autoantibody Reactivity and Corresponding Protein Levels.
The statistical methods (a) included analysis of variance (ANOVA) to take into account variation from multiple effects for each protein, filtering of interaction effects and post hoc Bonferroni correction to arrive at a set of three proteins, AKT3, FCGR3A and ARL8B. The box plots (b) illustrate the differences between the MDS patients and the healthy cohort for autoantibody reactivity to each protein of interest, with corresponding protein levels assessed by ELISA assays shown in (c). [The thickness of each box plot reflects the corresponding subject numbers (112 Healthy, 119 s-MDS, 42 t-MDS, 43 L) – see also Table 1b].
To classify the different samples into patient subgroups we implemented Kernel Discriminant Analysis (KDA24,25,26), using the transformed intensities for AKT3, FCGR3A and ARL8B, for successful classification into retrospective classes, particularly for s-MDS (Fig. 4a–b). When considering s-MDS and healthy cohort only, s-MDS samples were classified correctly 87% of the time (103/119, standard deviation, s.d. ~3.1) and healthy samples at 90% (101/112, s.d. ~2.7). Expanding the classification to all MDS and L patients did not significantly affect classification for s-MDS or healthy cohorts (Fig. 4b). However, the classification was lower for known t-MDS (38%, 16/42, s.d. ~3.0) and L (30%, 13/43, s.d. ~2.3) samples. Autoantibody reactivity-based classification worked less well for classifying samples into their known IPSS classes (Fig. 4c). The best classification was for Intermediate 1(62%, 46/74 s.d, ~3.2), while the classification of healthy samples was at 100% (112/112, s.d ~0.6).
Classification of MDS Patients Using Autoantibody Reactivity Levels.
The standardized reactivity of each of the three proteins of interest was used as a three-dimensional coordinate to classify patient subgroups. (a) Classification into retrospective s-MDS and healthy subsets was the most successful. The example figure indicates the original data points (green: healthy samples, blue: s-MDS), superimposed on the corresponding classification results, kernel density estimates, as computed using Kernel Discriminant Analysis (KDA). Introducing all retrospective categories (b) did not affect s-MDS or healthy classification, though t-MDS and AML displayed lower classification based on the protein of interests. Subject total numbers are as indicated for Stage II in Table 1. The actual data points are superimposed over the densities computed by the KDA for all MDS subgroups, showing more overlap than in (a). (c) Equivalent classification into IPSS risk groups does not perform as well as classification into retrospective subsets. (d) All shown classifications, (a–c), were performed by KDA, using AKT3, FCGR3A and ARL8 standardized autoantibody reactivities to represent three coordinates, for assigning each sample to a point in a three-dimensional space. The classification involved 5-fold cross-validation and 1,000 repetitions for each data partitioning to assess variance and median classification (see also Supplementary Tables S4–S6). N.B. The above classification matrices display classification medians, which might lead to lower sums due to rounding, mismatching the total sum of samples used in each row.
We detected all three proteins of interest in plasma using Enzyme-linked Immunosorbent Assays (ELISAs). For AKT3 and FCGR3A the differential protein levels were found to be statistically significant (ANOVA analysis Bonferroni corrected P < 9 × 10−8, <7 × 10−5 respectively) (Fig. 3c). For AKT3 the differential protein trend between retrospective classes reflected the autoantibody reactivity (post hoc tests, Bonferroni P < 0.01, displaying differences between all patient groups versus healthy, as well as L versus t-MDS). For FCGR3A the differential protein levels showed an opposite trend compared to the corresponding autoantibody reactivity, Fig. 3b–c (post hoc tests, Bonferroni p < 0.01 displaying differences between healthy versus both t-MDS and s-MDS, as well as between L and both t-MDS and s-MDS).
Discussion
Our investigation demonstrated differential autoantibody reactivity in MDS patient subsets, which was distinct from healthy individuals. The autoantibodies displaying increased protein reactivity compared to healthy patients included several unique proteins for t-MDS, s-MDS and L in both the initial and validation stages (Stage I and II) of our investigation. The finding that increased antibody reactivity for these proteins predominantly occurred in s-MDS patients is consistent with prior data indicating early stage MDS having a higher degree of immune-related abnormalities than later stage patients16,17. These proteins are involved in cancer-relevant biological processes such as apoptosis and have associations involving autoimmune responses. Differential reactivity of these antibodies between early stage MDS and healthy individuals also has diagnostic utility since such clinical distinctions morphologically are often difficult27.
The three proteins validated in Stage II of our investigation present a more robust set, showing increased autoantibody reactivity in MDS patients. The detected autoantibody reactivity to the above proteins offers possible new connections between immune response and tumor formation and detection. Of these, we particularly note that AKT3 (v-akt murine thymoma viral oncogene homolog 3 /protein kinase B, gamma), has been known to be actively involved in multiple cancers, including breast cancer28,29, tumorigenesis in melanomas30,31, lung cancers32 and ovarian cancers33 (cell cycle involvement). In previous studies AKT3 gene overexpression and deregulation of AKT pathways, have been detected by gene expression profiling23,34. In addition, our observation of differential autoantibody reactivity and corresponding protein levels, make this a compelling case for the involvement of AKT3 in MDS and highlight its potential as a biomarker for the condition in s-MDS. FCGR3A (an IgG receptor) is involved in multiple antibody processes. Fc-receptors have been the target of multiple therapeutic approaches for cancer and inflammatory states35, including FCGR3A36 (genotype association to rituximab response). The FCGR3A protein levels as determined by ELISA showed an opposite trend as compared to the corresponding autoantibody reactivity. While the FCGR3A protein levels may reflect the possibility that the observed autoantibody reactivity is due to “non-specific” Fc-Receptor interaction rather than increased amount of specific anti-FCGR3A antibody, it does not necessarily imply a correlation between the amount of antigen and the strength of reactivity. In fact, many highly expressed proteins are not autogenic (e.g. albumin, which is a negative acute-phase protein with decreased plasma levels during immune response37,38,39), while a small amount of allergen could trigger severe immune response. ARL8B (ADP-ribosylation factor-like 8B) is involved in lysosome trafficking and positioning, influencing mTOR expression (mammalian target of rapamycin) and autophagosome formation40 and may also be associated to metastasis and tumorigenesis41. Patient classification may be made into two separate classification schemes, prospectively by IPSS or retrospectively (s/t/L or healthy) as considered above. These classification schemes show good overlap as the IPSS Low and Intermediate 1 risk categories account for 27% and 68% respectively of our s-MDS patients (85% total). Using the autoantibody reactivities we classified patients into each of these two classification schemes (retrospective or IPSS), showing better concordance of autoantibody reactivity in stable patients with the retrospective classification rather than IPSS, but slightly lower with healthy patients (90%). As the classification into IPSS does better on healthy patients (100%), this suggests that combining the IPSS classification scheme with the autoantibody reactivities may improve clinical diagnosis of s-MDS patients while maintaining low numbers of false positives in healthy individuals. Furthermore, autoantibody reactivity-based classification relies on direct molecular marker signatures and may have possible application as an alternative or supplement to the IPSS risk estimation, especially for early detection of s-MDS patients. Finally, the detection of autoantibody reactivity would only require plasma and be minimally invasive for the patients.
Our study discovered differential autoantibody reactivity in MDS and identified autoantibodies particular to prognostic MDS subsets, demonstrating that protein microarrays provide a powerful approach to identify unique biomarkers associated with this disease. In our investigation we chose the more sensitive focused protein array approach over traditional chromogenic ELISA for our validation purposes. While we envision that high-throughput, fluorescence-based methods such as protein microarrays may eventually replace traditional chromogenic ELISA in clinical tests, we expect ELISA clinical tests may also be developed in the future based on our findings. Regarding the possible specificity of our findings, although such autoantibody studies have not as yet been reported in other hematologic malignancies, the combined autoantibody reactivity we observed in our MDS patients was distinct from those reported in patients with ovarian13 and prostate14 cancer, diabetes11 and after respiratory infections10. We expect our findings of the increased autoantibody reactivity in MDS patients with relatively prolonged clinical courses will encourage future studies with larger patient cohorts, that will be helpful to further substantiate the prognostic importance of such data and exploration of the underlying molecular mechanisms. The identified autoantibody reactivity may greatly enhance our diagnostic and prognostic capabilities for MDS.
Methods
MDS patients
Patient recruitment and all plasma sample collections were carried out at Stanford University Medical Center with informed consent, under approval of the Stanford Internal Review Board for Human Subjects. The plasma samples were randomly obtained from a group of MDS patients (seen 2001–2010), for whom long-term followup was available. Samples were obtained at either the time of MDS diagnosis or during their disease course [median 3 months from diagnosis (range 0–99), mean +/− SEM 11.5 +/− 1.5 months].
Plasma autoantibody reactivity profiling with protein arrays
In Stage I the autoantibody reactivity was profiled on the retrospective patient subsets (Table 1a) using Invitrogen ProtoArray Protein Microarrays, v5.0 (9,483 unique human proteins spotted in duplicate, 23,232 signals total including controls), as described previously12,13. In parallel, duplicate negative control arrays were probed. The plasma samples were diluted 1:100 in 5 ml Washing Buffer (1× PBS, 0.1% Tween 20, 1× Roti-Block). The ProtoArrays were dried and scanned using a Genepix 4200AL Microarray Scanner (Molecular Devices, Sunnyvale, CA). The obtained array images were analyzed (Genepix Pro 6.1, Molecular Devices), obtaining feature locations, signal foreground and background intensity quantification and corresponding identification information (.gpr file format). In addition to probing for IgG reactivity (532 nm channel), the arrays were spotted with proteins containing a glutathione S-transferase (GST) fusion tag and a second channel (635 nm) screening allowed us to probe GST intensity, which is used as a proxy for protein concentration per array spot (see also Supplementary Fig. S2).
In Stage II a custom protein array was created to probe autoantibody reactivity in an enlarged cohort (Table 1b, Figure 2a). Each array was produced through Invitrogen, using ProtoArray technology (Figure 2b–c) and comprised of 12 blocks, [176 duplicated protein spots (total 352) per block]. 150 proteins from Stage I were used in analyzing protein reactivity (one subject sample per block), including 25 of the differentially reacting proteins of interest identified in Stage I and 125 randomly selected control proteins. The probing procedure was carried out as in Stage I described above, including a negative control. The array images were analyzed as in Stage I to obtain signal information, with each block considered separately, including probing for GST intensity.
Array analyses, stage I
The arrays were analyzed in a multi-step process, involving inter- and intra-array normalization and signal comparisons between groups to identify initially a high-interest protein set that displayed increased IgG reactivity in patient subgroups (Fig. 1a). For each array, per channel intra-array normalization was performed via implementing the ProCAT algorithm42 (with sliding window parameter length 15), which takes into account local background subtraction and local intensity normalization across each array. This local intensity adjustment is necessary to correct for array variations stemming from the printing and probing procedures. To adjust for probing and scanning procedure variations, inter-array intensities were quantile normalized43. The probed intensities were compared between all binary comparisons of patient retrospective and healthy groups. The procedure has been previously shown to be highly reproducible (R2 > 0.89 for both technical replicates and duplicated spots12).
In our study we compared different patient subgroups to one another. We were specifically interested in signals showing increased reactivity in the retrospectively analyzed MDS patients versus the healthy control group, particularly s-MDS and assessed statistical significance through a two-tailed Mann-Whitney test (Mathematica 9.0; multiple hypotheses Bonferroni adjusted P < 0.01).
After filtering control signals and protein spots inconsistently printed on the array (i.e. displaying differential GST-tag signals due to variability in array printing), a total of 35 proteins showed increased reactivity at a cutoff of P < 4 × 10−7 (Bonferroni adjusted P < 0.01) and were identified as high-interest proteins from comparisons of MDS subsets to the healthy cohort (Fig. 1b–c). These were used for pathway and functional analysis through IPA (Ingenuity® Systems, http://www.ingenuity.com), to identify relevant biological functions in the Ingenuity Knowledge Base (Fig. 1d). To determine the probability that each biological function assigned to the data set was due to chance alone, P values were calculated using right-tailed Fisher's exact tests (P < 0.05). Additionally, canonical pathways from the IPA library that were most statistically significant were ascertained based on p-value (p < 0.05, Fisher's exact test) and ratio of molecules from the data set as compared to the total in the network (see also Supplementary Information).
Array analyses, stage II
Subsequently, in the validation stage of the study we successfully expressed 25 of these proteins onto customized focused arrays as described above. We note here that 10 proteins from the initial list of 35 candidates were not included in the validation because these could either not be successfully expressed, isolated or printed (lack of the proper clone; low viability for a specific clone; low yield in isolation, printing error, etc.) or did not pass our strict secondary quality control during selection for focused array printing, as an added measure for validation reproducibility (large GST-signal variation, negative flags raised in scanning, saturation of signal and non-reproducible variance stability in signal analysis). Additionally, the study was expanded to a much larger cohort (Table 1b). The protein arrays were scanned as described above, with each block corresponding to a different patient and patient samples performed in duplicates. We used the same inter- and intra-array normalization as in Stage I (Fig. 2a), applied per block and the signals for each protein spotted were transformed to a normal distribution using a Box-Cox transformation44. Each protein was analyzed using an ANOVA model45, checking for variance in subject subgroup (retrospective classification), replication (duplicate samples, duplicate protein spots), subject age, gender, marrow blast levels (healthy, IPSS blast categories, leukemic status) and randomization effects (batch effects between Stage I or II of the investigation), Fig. 3a. The possible interactions between all the above were also considered. Signals showing significant primary effects but no significant interactions, using Bonferroni post-hoc tests to correct for multiple group and feature comparisons, were selected as high–confidence signals of interest. Additionally, the ANOVA was used to screen out signals showing significant effects due to replication, subject age and randomization. Furthermore, to ensure that protein levels were not varying between arrays, statistically significant differential GST signals were eliminated.
Based on the ANOVA, standardized transformed reactivity levels of AKT3, ARL8B, FCGR3A were found to be higher in patients versus healthy cohort (Fig. 3b). These proteins were then used for classifications, namely KDA (R46 package ks47, using unconstrained smoothed cross-validation method for bandwidth selection) and Linear Discriminant analysis (LDA; R46 package MASS48, see Supplementary Tables S4–S6). KDA outperformed LDA for retrospective and IPSS classifications. Both methods involved assigning classes with 5-fold cross-validation and performing 1,000 random data partitions (Fig. 4).
ELISAs
Utilizing ELISA kits we measured plasma levels of AKT3 (dilution 1:5, PathScan® Catalog No. #7934, Cell Signaling), FCGR3A (dilution 1:10, Catalog No. E91278Hu, Uscn Life Science Inc.) and ARL8B (dilution 1:10, MyBioSource Catalog No. MBS946943), using samples from 20 subjects (80 total) for each of s-MDS, t-MDS, L and healthy subjects tested in triplicate, following each manufacturer's instructions (Fig. 3c).
Additional Information
Accession codes: Protein array data and custom array design were submitted to Gene Expression Omnibus (GEO) repository under accessions GSE48155 and GPL17321 respectively.
References
Greenberg, P. L. Current therapeutic approaches for patients with myelodysplastic syndromes. Br. J. Haematol. 150, 131–143, 10.1111/j.1365-2141.2010.08226.x (2010).
Tefferi, A. & Vardiman, J. W. Myelodysplastic syndromes. N. Engl. J. Med. 361, 1872–1885, 10.1056/NEJMra0902908 (2009).
Raza, A. et al. Simultaneous assessment of cell kinetics and programmed cell death in bone marrow biopsies of myelodysplastics reveals extensive apoptosis as the probable basis for ineffective hematopoiesis. Am. J. Hematol. 48, 143–154, 10.1002/ajh.2830480302 (1995).
Rajapaksa, R., Ginzton, N., Rott, L. S. & Greenberg, P. L. Altered oncoprotein expression and apoptosis in myelodysplastic syndrome marrow cells. Blood 88, 4275–4287 (1996).
Parker, J. E. et al. The role of apoptosis, proliferation and the Bcl-2-related proteins in the myelodysplastic syndromes and acute myeloid leukemia secondary to MDS. Blood 96, 3932–3938 (2000).
Greenberg, P. L. Molecular and genetic features of myelodysplastic syndromes. Int. J. Lab. Hematol. 34, 215–222, 10.1111/j.1751-553X.2011.01390.x (2012).
Bejar, R. et al. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 364, 2496–2506, 10.1056/NEJMoa1013343 (2011).
Greenberg, P. et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 89, 2079–2088 (1997).
Greenberg, P. L. et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 120, 2454–2465, 10.1182/blood-2012-03-420489 (2012).
Zhu, H. et al. Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc. Natl. Acad. Sci. U. S. A. 103, 4011–4016, 10.1073/pnas.0510921103 (2006).
Winer, D. A. et al. B cells promote insulin resistance through modulation of T cells and production of pathogenic IgG antibodies. Nat. Med. 17, 610–617, 10.1038/nm.2353 (2011).
Chen, R. et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148, 1293–1307, 10.1016/j.cell.2012.02.009 (2012).
Hudson, M. E., Pozdnyakova, I., Haines, K., Mor, G. & Snyder, M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc. Natl. Acad. Sci. U. S. A. 104, 17494–17499, 10.1073/pnas.0708572104 (2007).
Wang, X. et al. Autoantibody signatures in prostate cancer. N. Engl. J. Med. 353, 1224–1235, 10.1056/NEJMoa051931 (2005).
Hamblin, T. Immunologic abnormalities in myelodysplastic syndromes. Hematol. Oncol. Clin. North Am. 6, 571–586 (1992).
Kordasti, S. Y. et al. IL-17-producing CD4(+) T cells, pro-inflammatory cytokines and apoptosis are increased in low risk myelodysplastic syndrome. Br. J. Haematol. 145, 64–72, 10.1111/j.1365-2141.2009.07593.x (2009).
Kordasti, S. Y. et al. CD4 + CD25high Foxp3+ regulatory T cells in myelodysplastic syndrome (MDS). Blood 110, 847–850, 10.1182/blood-2007-01-067546 (2007).
Richert-Boe, K. E. & Bagby, G. C., Jr In vitro hematopoiesis in myelodysplasia: liquid and soft-gel culture studies. Hematol. Oncol. Clin. North Am. 6, 543–556 (1992).
Ohyashiki, J. H. et al. Telomere shortening associated with disease evolution patterns in myelodysplastic syndromes. Cancer Res. 54, 3557–3560 (1994).
Sashida, G. et al. Telomere dynamics in myelodysplastic syndrome determined by telomere measurement of marrow metaphases. Clin. Cancer Res. 9, 1489–1496 (2003).
Jiang, H. et al. Proteins induced by telomere dysfunction and DNA damage represent biomarkers of human aging and disease. Proc. Natl. Acad. Sci. U. S. A. 105, 11299–11304, 10.1073/pnas.0801457105 (2008).
Zhong, L. et al. Protein microarray: sensitive and effective immunodetection for drug residues. BMC Biotechnol. 10, 12, 10.1186/1472-6750-10-12 (2010).
Sridhar, K., Ross, D. T., Tibshirani, R., Butte, A. J. & Greenberg, P. L. Relationship of differential gene expression profiles in CD34+ myelodysplastic syndrome marrow cells to disease subtype and progression. Blood 114, 4847–4858, 10.1182/blood-2009-08-236422 (2009).
Mika, S., Ratsch, G., Weston, J., Scholkopf, B. & Mullers, K. Fisher discriminant analysis with kernels. Neural Networks for Signal Processing IX, 1999. Proceedings of the 1999 IEEE Signal Processing Society Workshop. 41–48, 10.1109/NNSP.1999.788121 (1999).
Baudat, G. & Anouar, F. E. Generalized discriminant analysis using a kernel approach. Neural Comput. 12, 2385–2404, 10.1162/089976600300014980 (2000).
Li, Y. M., Gong, S. G. & Liddell, H. Recognising trajectories of facial identities using kernel discriminant analysis. Image Vision Comput. 21, 1077–1086, 10.1016/J.Imavis.2003.08.01 (2003).
Font, P. et al. Inter-observer variance with the diagnosis of myelodysplastic syndromes (MDS) following the 2008 WHO classification. Ann. Hematol. 92, 19–24, 10.1007/s00277-012-1565-4 (2013).
Banerji, S. et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486, 405–409, 10.1038/nature11154 (2012).
Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70, 10.1038/nature11412 (2012).
Stahl, J. M. et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 64, 7002–7010, 10.1158/0008-5472.CAN-04-1399 (2004).
Davies, M. A. et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br. J. Cancer 99, 1265–1268, 10.1038/sj.bjc.6604637 (2008).
Cancer Genome Atlas Research, N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525, 10.1038/nature11404 (2012).
Cristiano, B. E. et al. A specific role for AKT3 in the genesis of ovarian cancer through modulation of G(2)-M phase transition. Cancer Res. 66, 11718–11725, 10.1158/0008-5472.CAN-06-1968 (2006).
Pellagatti, A. et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia 24, 756–764, 10.1038/leu.2010.31 (2010).
Hogarth, P. M. & Pietersz, G. A. Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat. Rev. Drug Discov. 11, 311–331, 10.1038/nrd2909 (2012).
Cartron, G. et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 99, 754–758 (2002).
Gabay, C. & Kushner, I. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 340, 448–454, 10.1056/NEJM199902113400607 (1999).
Gruys, E., Toussaint, M. J., Niewold, T. A. & Koopmans, S. J. Acute phase reaction and acute phase proteins. J. Zhejiang Univ. Sci. B 6, 1045–1056, 10.1631/jzus.2005.B1045 (2005).
Ritchie, R. F. et al. Reference distributions for the negative acute-phase serum proteins, albumin, transferrin and transthyretin: a practical, simple and clinically relevant approach in a large cohort. J. Clin. Lab. Anal. 13, 273–279 (1999).
Korolchuk, V. I. et al. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453–460, 10.1038/ncb2204 (2011).
Mohamed, M. M. & Sloane, B. F. Cysteine cathepsins: multifunctional enzymes in cancer. Nat. Rev. Cancer 6, 764–775, 10.1038/nrc1949 (2006).
Zhu, X., Gerstein, M. & Snyder, M. ProCAT: a data analysis approach for protein microarrays. Genome Biol. 7, R110, 10.1186/gb-2006-7-11-r110 (2006).
Bolstad, B. M., Irizarry, R. A., Astrand, M. & Speed, T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003).
Box, G. E. & Cox, D. R. An analysis of transformations. J. R. Stat. Soc. Ser. B Stat. Methodol. 26, 211–252 (1964).
Pavlidis, P. Using ANOVA for gene selection from microarray studies of the nervous system. Methods 31, 282–289 (2003).
R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2013).
Duong, T. ks: Kernel density estimation and kernel discriminant analysis for multivariate data in R. J. Stat. Softw. 21, 1–16 (2007).
Venables, W. N., Ripley, B. D. & Venables, W. Modern applied statistics with S-PLUS. Vol. 250 (Springer-Verlag New York, 1994).
Acknowledgements
G.I.M.'s research reported in this publication is supported by the National Human Genome Research Institute (NHGRI) of the National Institutes of Health under Award Number K99HG007065 and previously T32HG000044. M.P.S.'s research was supported by NHGRI under Award Number P50HG002357. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. P.L.G.'s research was supported by the Stanford MDS Research Fund, the William E. Walsh Leukemia Research Fund, the Eugene, Elizabeth and Christina Cronkite Fund for Hematology, the Leukemia & Lymphoma Society (SCOR grant) and the Veterans Administration Palo Alto Health Care System (resources and use of facilities).
Author information
Authors and Affiliations
Contributions
G.I.M. and R.C. contributed equally to the project, conceived the study, project planning, did experimental design and work and statistical analysis and wrote the manuscript. Y.Z., K.S., D.S. and H.I. were involved in experimental work, manuscript editing; L.X. was involved in financing and supervision of Y.Z. and manuscript editing, P.L.G. and M.P.S. supervised the project and were involved in project planning, financing, supervision and manuscript preparation. P.L.G. and M.P.S. are both senior authors.
Ethics declarations
Competing interests
M.P.S. serves as founder and consultant for Personalis, is a member of the scientific advisory board of GenapSys and a consultant for Illumina.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Mias, G., Chen, R., Zhang, Y. et al. Specific Plasma Autoantibody Reactivity in Myelodysplastic Syndromes. Sci Rep 3, 3311 (2013). https://doi.org/10.1038/srep03311
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03311
This article is cited by
-
Myelodysplastic syndrome in an infant with constitutional pure duplication 1q41-qter
Human Genome Variation (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.