Abstract
Ribosomes contain a number of modifications in rRNA, the function of which is unclear. Here we show – using proteomic analysis and dual fluorescence reporter in vivo assays – that m2G966 and m5C967 in 16S rRNA of Escherichia coli ribosomes are necessary for correct attenuation of tryptophan (trp) operon. Expression of trp operon is upregulated in the strain where RsmD and RsmB methyltransferases were deleted, which results in the lack of m2G966 and m5C967 modifications. The upregulation requires the trpL attenuator, but is independent of the promotor of trp operon, ribosome binding site of the trpE gene, which follows trp attenuator and even Trp codons in the trpL sequence. Suboptimal translation initiation efficiency in the rsmB/rsmD knockout strain is likely to cause a delay in translation relative to transcription which causes misregulation of attenuation control of trp operon.
Similar content being viewed by others
Introduction
The ribosome is a 2.7-MDa machine that is composed of proteins and ribosomal RNAs (rRNA). Although the main function of the ribosome is to synthesize proteins, an equally important function is to maintain translation efficiency of different mRNA species precisely as needed for the overall cell fitness at particular environmental conditions. Ribosomes contain a number of ubiquitous rRNA modifications, e.g. 24 methylated nucleotides in rRNA from E. coli1. Recently a list of genes responsible for methylation of rRNA has been completed2. All modified nucleosides in rRNA are located in the functional centers of the ribosome3. Especially rich in modifications is the P site of the ribosome; however, the functional role of majority of modified nucleotides remains obscure1,4. Not a single gene coding for rRNA modification enzyme appeared to be essential for viability at the standard laboratory conditions of E. coli cultivation. It became a common point to suggest that modified nucleotides might be essential at some conditions which are rarely tested in the laboratory1. Thus, although modification of rRNA might be dispensable for all the basic steps of translation, it is possible that rRNA modification is needed for the control over translational efficiencies of some mRNAs at particular conditions.
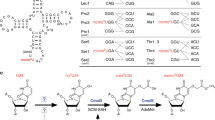
Nucleosides m2G966 and m5C967 of 16S rRNA are located at the P site of the 30S subunit in direct contact with the anticodon of the tRNA. Although there are no direct evidence on the regulation of the corresponding rRNA methyltransferases RsmD and RsmB, analysis of co-expression data suggests that at least rsmB gene expression is correlated with that of a set of genes related to translation4. Previously we described ΔrsmB/ΔrsmD strain lacking RsmD and RsmB methyltransferases and hence devoid of the two methyl groups, which had a cold-sensitive phenotype and reduced fitness when compared with the wild-type parent strain5. The ribosomes purified from the ΔrsmB/ΔrsmD strain had a mild kinetic defect in the selection of initiator fMet-tRNA in certain mRNA contexts, suggesting that the methylations might have a differential effect on translation initiation of a subset of cellular mRNAs5. A role for the modified nucleosides m2G966 and m5C967 of 16S rRNA in initiation was further supported by recent findings using specialized reporter system based on mutant initiator tRNA6. Here we tested impact of m2G966 and m5C967 modification on the global proteome of E. coli and analyzed in detail the effect of upregulation of the trp operon in the strain lacking G966 and C967 modifications.
Tryptophan operon is a textbook example of the gene expression regulation based on transcription attenuation mechanism7,8. The attenuation of trp operon entails pausing of ribosomes translating operon leader region trpL, on the tandem Trp-coding UGG codons7. Such pausing is normally caused by Trp-tRNATrp deficiency during Trp starvation and leads to the formation of the transcript secondary structure inhibiting premature transcript termination. In contrast, efficient translation of UGG codons by a ribosome leads to the formation of the transcript secondary structure, which causes termination of transcription.
In this work we analysed how the lack of two 16S rRNA modifications caused misregulation of trp operon.
Results
Comparative proteome analysis of the ΔrsmB/ΔrsmD strain
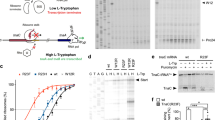
To identify the proteins which were differentially expressed depending on the lack of G966/C967 methylation we investigated the proteome of ΔrsmB/ΔrsmD strain in rich LB (Figure 1a) and poor M9 (Figure 1b) media at the logarithmic phase and in the LB media at the stationary phase (Figure 1c). The wild type proteome was labeled with Cy3 green fluorescent dye, while the proteome of the ΔrsmB/ΔrsmD strain was labeled with Cy5 red fluorescent dye. Fluorescently labeled total protein samples were mixed to equal Cy3 and Cy5 total fluorescence and subjected to 2D protein gel separation. Protein spots whose Cy5/Cy3 fluorescence ratio was below 0.5 or above 2 were considered significantly under- or over-represented in the proteome of the mutant bacteria (Supplementary file 2) and the proteins analyzed by MALDI-MS analysis after tryptic digestion9. Additionally, the wild type and ΔrsmB/ΔrsmD strains were compared by LC/MS of the total proteome tryptic digest, so called shotgun proteome analysis (Supplementary file 3). Distortions in the proteome observed by both methods were considered highly reliable.
Comparison of the wild type and ΔrsmD/ΔrsmB strain proteomes.
2D gel electrophoresis of Cy3-labeled proteome from the wild-type strain and Cy5-labeled proteome from ΔrsmD/ΔrsmB strain. Yellow spots correspond to the proteins with unchanged expression level. Green spots reflect proteins underrepresented in ΔrsmD/ΔrsmB strain. Red spots correspond to proteins overrepresented in ΔrsmD/ΔrsmB strain. The spots analyzed by MALDI MS analysis after trypic digestion are depicted by arrows. Cells were grown in the rich LB media to (a) logarithmic stage, (b) in poor M9 medium to logarithmic stage, or (c) in LB medium to the stationary phase.
Most of differences in the protein composition of the wild type and ΔrsmB/ΔrsmD strains were observed at the logarithmic growth phase (Figure 1a, b), whereas at the stationary phase protein compositions of the mutant and wild type strains were similar (Figure 1c). We noted the lack of rRNA methylations altered the abundance of several proteins the synthesis of which is regulated by the transcription attenuation mechanism: e.g. translation of trpB, encoded within the trpLEDCBA (trp) operon, was derepressed in the ΔrsmB/ΔrsmD strain even in the presence of tryptophan (Trp) (Supplementary file 2), while translation of pheS coded by pheMST operon was downregulated (Supplementary file 2). Upregulation of expression of the trp operon in the ΔrsmB/ΔrsmD strain was evidenced by two independent methods of comparative proteome analysis. For pheMST operon downregulation was observed (Supplementary file 2). Such changes in protein abundance might be directly caused by the lack of methylations or indirectly through global regulation mechanisms which are sometimes difficult to trace. In the following we investigated whether the lack of m2G966 and m5C967 methylations have a direct effect on the regulation of trp operon.
Promoter activity vs. attenuation of trp operon
Trp operon is regulated by two distinct mechanisms: (i) by transcription attenuation which is relieved by the ribosome pausing during translation of trpL leader gene7 and (ii) by Trp-stimulated binding of the transcription repressor TrpR to the promoter of trp operon10. We first checked the second possibility, i.e. whether the promoter plays a role in the upregulation of trp operon in ΔrsmB/ΔrsmD strain. We used a reporter system based on dual fluorescence assay which was recently developed in our laboratory11,12. The plasmid pRFPCER (Figure 2a) encoded red (RFP) and cerulean (CER) fluorescent proteins which have spectral properties allowing for their simultaneous detection in the same bacterial culture. RFP gene, which was cloned after the phage T5 promoter, was used as internal control, while trp operon promoter was inserted upstream of CER (Figure 2b, left panel). Expression of CER protein under the control of trp promoter did not depend on G966/C967 methylations (Figure 2b, right panel), suggesting that the upregulation of trp operon in the ΔrsmB/ΔrsmD strain cannot be explained by altered activity of trp promotor. The result also suggests that intracellular tryptophan concentration was not affected by rsmB and rsmD inactivation, which would otherwise lead to an altered efficiency of trp promoter.
Role of m2G966/m5C967 in attenuation of expression of trp operon.
Drawn are CER/RFP expression ratios (a–j) or Fluc/Rluc ratio (k). Green bars show expression in the wild type strain, red bars in ΔrsmB/ΔrsmD double knockout strain. (a) Parent construct pRFPCER; (b) pRFPCER-Ptrp; (c) pRFPCER-TrpL; (d) pRFPCER-TrpL2A; (e) pRFPCER-TrpL2R; (f) pRFPCER-TrpE; (g) pRFPCER-TrpL2A-standard rbs; (h) pRFPCER-HisL; (i) pRFPCER-PheM; (j) pRFPCER-IlvL; (k) pLuc-TrpL.
Altered attenuation of trp operon in ΔrsmB/ΔrsmD strain
To test the effect on attenuation, we fused the sequence coding for trp operon leader, trpL, together with its natural translation initiation region and ten N-terminal amino acids of the first structural gene in trp operon (trpE), to the N-terminus of the CER gene; RFP was used as an internal control. Previous experiments validated that the reporter construct was sensitive to Trp concentration12. Both reporter genes were transcribed from identical T5 phage promoters to avoid an interference of transcription initiation with the expression of reporter constructs. As expected, the reporter was induced in the ΔrsmB/ΔrsmD strain (Figure 2c), in agreement with the results of comparative proteome analysis; the induction was not due to changes in the efficiency of trp promoter, because T5 phage promoter was used. Thus, upregulation of trp operon in ΔrsmB/ΔrsmD strain is caused by improper function of the attenuation mechanism.
To further check if observed upregulation of the gene following trp attenuator is not specific to promoter or particular reporter gene system we created a dual luciferase reporter construct where both Renilla and Firefly luciferase genes were placed under a control of identical Para promoters. Additionally, in front of Firefly luciferase we inserted trp attenuator region, while Renilla luciferase was used as internal control (Figure 2k, left panel). After transformation of the wild type and ΔrsmB/ΔrsmD strains with this reporter construct and induction of the promoters with arabinose, we measured luciferase activities. We observed an increase in expression of the reporter Firefly luciferase gene following trp attenuator region in the strain deficient in G966/C967 methylations (Figure 2k, right panel; compare green bar (WT) with red bar (ΔrsmB/ΔrsmD)) in complete agreement with our earlier observations obtained by proteomics and dual fluorescent protein reporter.
Trp codons are dispensable for trp operon upregulation caused by lack of the 16S rRNA nucleotides G966/C967 methylation
Natural regulation of trp operon depends on stalling of translating ribosome at two Trp codons in trpL leader upon Trp starvation. To check the role of the tandem Trp codons, we replaced the two UGG codons in the trpL gene with two GCG codons coding for Ala; the resulting reporter was not sensitive to tryptophan concentration12. A compensatory change was made to maintain the mRNA secondary structure of the attenuator region as described12. Substitution of Trp codons by efficiently translated alanine codons favors repression of the following gene expression, since it mimic the condition of tryptophan abundance. Substitution of Trp codons by Ala codons have not abolished upregulation of attenuation-dependent reporter gene in the strain lacking G966/C967 methylations (Figure 2d). Substitution of Trp codons with the rare Arg codons resulted in permanent derepression of reporter since it imitates tryptophan starvation. Derepression caused by rare Arg codons is independent on the 16S rRNA modification (Figure 2e) presumably because it corresponds to the maximal extent of transcription read through the attenuator. To further prove that upregulation of trp operon in the ΔrsmB/ΔrsmD strain demonstrated with the help of CER reporter was not due to the difference in ribosome binding sites, we created two more reporter constructs (Figure 2f–g). Deletion of trp operon attenuator region while retaining the ribosome binding site of CER gene same as for trpE gene completely abolished upregulation caused by lack of G966 and C967 methylation (Figure 2f). In contrast, replacement of trpE ribosome binding site ahead of CER by one unrelated to trp operon (Figure 2g) while preserving attenuator region as in the construct having double alanine codons (Figure 2d) do not prevent reporter upregulation by the lack of G966 and C967 methylation. Thus, not any particular feature of ribosome binding site of the trpE gene, but attenuator structure itself is responsible for the observed effect.
Lack of G966/C967 modification moderately downregulates phe and his operons
Several operons related to amino acids biosynthesis and tRNA aminoacylation are regulated by attenuation mechanism13,14,15. We tested whether attenuators of hisLGDCBHAFI, pheMST and ilvLGMEDA operons are sensitive to the lack of G966/C967 methylation. Parts of operons from the transcription start to the coding part of the second gene were cloned between the T5 promoter and the coding part of CER reporter gene; again, RFP gene was used as a control. The expression of the reporters in the wild type and ΔrsmB/ΔrsmD strains demonstrated a moderate repression of phe and his attenuators by the lack of G966/C967 methylations (Figure 2h–j).
Discussion
In this work we have demonstrated that methylations of two residues in 16S rRNA in the vicinity of the P site affect regulation of trp operon by attenuation in vivo. Although the ΔrsmB/ΔrsmD cells were cold-sensitive and had compromised fitness, there was almost no effect on the global translation elongation in vitro5 and in vivo16. The present results suggest that the shape of the proteome at exponential growth phase is altered in the ΔrsmB/ΔrsmD cells and that in particular expression of those genes is affected which are regulated by the attenuation mechanism. Trp operon upregulation in the strain devoid of G966 and C967 methylation in the 16S rRNA was highly reproducible and observed by the different proteomic methods in the independent laboratories and by two different reporter systems. Lack of influence of rRNA modification on trp operon expression at stationary phase could reflect overall decrease in translation efficiency and more prominent role of TrpR transcription repression, which is independent on translation.
Attenuation relies upon the interplay of simultaneous transcription and translation of the leader region of amino acids biosynthetic operons8. Efficient translation of the trpL region possible at the Trp abundance leads to the formation of transcription terminator (Figure 3a) at the time, when RNA polymerase passes this region. Stalling of the ribosome at the Trp coding UGG codons leads to the antiterminator secondary structure formation at the time RNA polymerase pass potential terminator (Figure 3b). Curiously, if translation is completely abolished, e.g. by a mutation in the start codon7 termination secondary structure is formed leading to the repression of further transcription (Figure 3c).
A model of the trp operon regulation by attenuation7,8 and the model for upregulation of trp operon in the strain devoid of modification of the 16S rRNA nucleotides m2G966/m5C967.
Ribosome is depicted as grey circle while RNA polymerase is shown as pink rectangle. Leader region of the trp operon is shown as a black line, while elements of attenuator region are shown in red and green. Also depicted are AUG start codon, UGG Trp-encoding codons and UGA stop codon. Each row depicts the sequence of events starting from the beginning of trp operon transcription (left to right). (a) Formation of the terminator structure in the case of Trp abundance. (b) Stalling of ribosome at UGG codons in the case of Trp starvation. Formation of antiterminator structure. (c) Formation of the anti-antiterminator structure in the case of complete absence of translation, as in start codon mutant described7. (d) A model of derepression of trp operon in the ΔrsmB/ΔrsmD double knockout strain. Delayed translation intiation leads to increasing the distance between RNA polymerase and ribosome. At the time RNA polymerase transcribes potential terminator, ribosome could only reach the beginning of attenuator region.
The effect of G966 and C967 methylations in 16S rRNA on the attenuation efficiency may be explained by a change is the subtle balance between the rates of transcription and translation leading to attenuation. Replacement of Trp codons with frequently used Ala codons lead to repression of the reporter CER gene following attenuator but did not abolish the constitutive upregulation of trp operon in the ΔrsmB/ΔrsmD strain. This may be a consequence of delay in translation initiation in the double knockout strain in agreement with previously published effect of G966 and C967 methylations in 16S rRNA on the translation initiation in vitro5 and in vivo6. Substitution of Trp codons with the rare Arg codons resulted in constitutive derepression of the gene following attenuator, indicating that ribosomes are stalled at rare codons similarly for the wild type strain and for the ΔrsmB/ΔrsmD strain. It is interesting to note a difference in response of trp, his and phe attenuators for the lack of G966 and C967 methylations in 16S rRNA. It might be explained by a difference in length and secondary structures of these attenuators. While the common mechanism of attenuation relieve is based on ribosome stalling at the starving codons, attenuation dependence on the translation initiation efficiency might be opposite, as was noted previously17,18.
Previous in vitro experiments indicated that ribosomes lacking modifications of G966 and C967 have a higher rate of fMet-tRNA dissociation from the 30S initiation complex prior to the start codon recognition5. Modification of G966 and C967 of the 16S rRNA does not have any influence on translation initiation at ribosome binding site of trpE gene. However, attenuator region of trp operon with Trp codons substituted with Ala codons granted derepression of the following CER reporter gene in the ΔrsmB/ΔrsmD strain even if CER gene possesses a ribosome binding site unrelated to trp operon. It is likely that in the strain lacking G966 and C967 nucleotide methylation, translation initiation is delayed at the trp leader due to higher probability of preinitiation complex dissociation (Figure 3d). This delay in translation allows RNA polymerase to proceed more towards the trp operon structural genes (see Figure 3d, middle panel). Widening of the gap between RNA polymerase and the ribosome mimic slower translation of the trpL reading frame similar to that observed at the tryptophan starvation. At the time RNA polymerase transcribes the potential terminator sequence, ribosome which started translation later due to lack of 16S rRNA modifications, could reach only the beginning of attenuator, making it possible to form the structure favorable for further transcription (Figure 3d).
The results of the present work provide an example for the role of rRNA modifications in regulation of gene expression in vivo. Such effects were previously demonstrated for induction of ermC expression19, another ribosome related mechanism of gene expression regulation in bacteria. In eukaryotes, changes in rRNA modification patterns were recently demonstrated for cancer cells as a consequence of p53 inactivation20. In turn, changes in rRNA modification lead to altered efficiency of cap- and IRES dependent translation in eukaryotes21. Thus, rRNA modifications – which themselves demand a large biosynthetic investment which may be controlled by the physiological status of the cell – might provide an additional, previously unrecognized mechanism of gene expression regulation and might be involved in the cell response to environmental changes.
Methods
E. coli strain BW2511322 was used as the wild type. Construction of ΔrsmB/ΔrsmD double knockout strain was described previously5. Proteomic analysis was performed as described9 (see also Supplementary file 1).
Reporter constructs containing RFP and CER genes (pRFPCER), TrpL (pRFPCER-TrpL) and mutant TrpL (pRFPCER-TrpL2A) were described previously12. Creation of other reporter constructs used in this work could be found in Supplementary file 1.
Chemically competent cells made from BW25113 strain and ΔrsmB/ΔrsmD double knockout strain were aliquoted (50 μl) into a 96-well plate by a Janus (Perkin Elmer) automated workstation and 1 μl of appropriate plasmid (1 ng) was added to each well. Next, the plate was incubated for 30 min at 4°C and after heat-shock (2 min at 44°C), 200 μl of LB medium was added to each well. After 1 hour of incubation at 37°C, 20 μl of transformation solution were transferred into the 96-well plate with LB-agar media, supplied with 100 μg/ml ampicillin; this transfer was repeated 3 times and three 96-well agar plates for three independent inoculations were obtained. Inoculations were produced by the Janus automated workstation and cells were grown overnight at 37°C in a 96-well 2 ml (Qiagen) plate with shaking (200 rpm). Cells were twice washed with 0.9% NaCl and the fluorescence of RFP and CER was measured by a Victor ×5 2030 (Perkin Elmer) multifunctional reader using appropriate emission/excitation filters (430/486 nm for CER and 531/595 nm for RFP). Standard deviation was derived from at least three parallel independent measurements.
Dual luciferase reporter assay23,24,25 was done according to the following procedure. Chemically competent cells made from BW25113 and ΔrsmB/ΔrsmD strains were transformed with reporter plasmid pLuc-TrpL. Cultures of individual clones were grown overnight in LB media supplemented with spectinomycin/streptomycin, diluted to 0.05–0.1 OD600 with fresh LB broth, incubated 2 hours with vigorous shaking before and 2 hours after induction with arabinose 1 mM. Cells were harvested by centrifugation 1 min 13000 rpm and the extracts were prepared by lysozyme-freezing method as described24. Extracts (10 μl) were taken for dual luciferase assay (Promega). Luminescence was measured using Luminometer 20/20n Turner Biosystems in relative light units (RLU). The activity of Renilla luciferase was used as internal control and the ratio of Firefly to Renilla luciferase activity was calculated. Standard deviation was derived from three parallel samples measurements obtained from three different colonies.
References
Sergiev, P. V. et al. Modifications of Ribosomal RNA: From Enzymes to Function. Ribosomes Rodnina, M. V., Wintermeyer, W. & Green, R. (eds.) 97–110 (Springer Vienna, 2011).
Golovina, A. Y. et al. The last rRNA methyltransferase of E. coli revealed: the yhiR gene encodes adenine-N6 methyltransferase specific for modification of A2030 of 23S ribosomal RNA. RNA 18, 1725–1734 (2012).
Brimacombe, R., Mitchell, P., Osswald, M., Stade, K. & Bochkariov, D. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J 7, 161–167 (1993)
Sergiev, P. V. et al. How much can we learn about the function of bacterial rRNA modification by mining large-scale experimental datasets? Nucleic Acids Res 40, 5694–5705 (2011).
Burakovsky, D. E. et al. Impact of methylations of m2G966/m5C967 in 16S rRNA on bacterial fitness and translation initiation. Nucleic Acids Res 40, 7885–7895 (2012).
Arora, S. et al. Distinctive contributions of the ribosomal P-site elements m2G966, m5C967 and the C-terminal tail of the S9 protein in the fidelity of initiation of translation in Escherichia coli. Nucleic Acids Res. 41, 4963–4975 (2013).
Zurawski, G., Elseviers, D., Stauffer, G. V. & Yanofsky, C. Translational control of transcription termination at the attenuator of the Escherichia coli tryptophan operon. Proc Natl Acad Sci USA 75, 5988–5992 (1978).
Merino, E. & Yanofsky, C. Transcription attenuation: a highly conserved regulatory strategy used by bacteria. Trends Genet 21, 260–264 (2005).
Sergiev, P. V. et al. Systems biology approach to the functional role of enzymatic modification of bacterial ribosome. Bioorg Khim 37, 81–90 (2011).
Gunsalus, R. P. & Yanofsky, C. Nucleotide sequence and expression of Escherichia coli trpR, the structural gene for the trp aporepressor. Proc Natl Acad Sci USA 77, 7117–7121 (1980).
Osterman, I. A., Evfratov, S. A., Sergiev, P. V. & Dontsova, O. A. Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res. 41, 474–486 (2013).
Osterman, I. A. et al. Attenuation-based dual-fluorescent-protein reporter for screening translation inhibitors. Antimicrob Agents Chemother 56, 1774–1783 (2012).
Barnes, W. M. DNA sequence from the histidine operon control region: seven histidine codons in a row. Proc Natl Acad Sci USA 75, 4281–4285 (1978).
Trudel, M. et al. Regulation of E. coli phenylalanyl-tRNA synthetase operon in vivo. Biochim Biophys Acta 782, 10–17 (1984).
Lawther, R. P. & Hatfield, G. W. Multivalent translational control of transcription termination at attenuator of ilvGEDA operon of Escherichia coli K-12. Proc Natl Acad Sci USA 77, 1862–1866 (1980).
Arora, S., Bhamidimarri, S. P., Weber, M. H. & Varshney, U. Role of the Ribosomal P-Site Elements of m2G966, m5C967 and the S9 C-Terminal Tail in Maintenance of the Reading Frame during Translational Elongation in Escherichia coli. J Bacteriol. 195, 3524–3530 (2013).
Mayaux, J. F. et al. Control of phenylalanyl-tRNA synthetase genetic expression. Site-directed mutagenesis of the pheS, T operon regulatory region in vitro. J Mol Biol. 184, 31–44 (1985).
Manabe, T. Theory of regulation by the attenuation mechanism: stochastic model for the attenuation fo the Escherichia coli tryptophan operon. J Theor Biol. 91, 527–544 (1981).
Vázquez-Laslop, N., Ramu, H., Klepacki, D., Kannan, K. & Mankin, A. S. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 29, 3108–3117 (2010).
Marcel, V. et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 24, 318–330 (2013).
Basu, A. et al. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular internal ribosome entry sites. Mol Cell Biol. 31, 4482–4499 (2011).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97, 6640–6645 (2000).
Grentzmann, G., Ingram, J. A., Kelly, P. J., Gesteland, R. F. & Atkins, J. F. A dual-luciferase reporter system for studying recoding signals. RNA 4, 479–486 (1998).
Kramer, E. B. & Farabaugh, P. J. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA 13, 87–96 (2007).
Harger, J. W. & Dinman, J. D. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 9, 1019–1024 (2003).
Acknowledgements
This work was supported by the Russian Foundation for Basic Research 11-04-01018 (AAB), 11-04-01314 (OAD) 12-04-33026-mol-a-ved (PVS), 13-04-00836-a (PVS), 14-04-01061 (OAD), 13-04-40211-H (AAB), 12-04-31132-mol-a (IAO), the Moscow University Development Program PNR 5.13 (OAD).
Author information
Authors and Affiliations
Contributions
I.A.O., M.V.S., I.A.D., M.A.G., O.V.P., I.A., S.K., D.G.A., V.M.G. carried proteomic analysis, I.V.P., I.A.O., D.E.B. performed dual reporter experiments, I.V.P., I.A.O., D.E.B., A.A.B. analyzed the data, P.V.S. and O.A.D. wrote the paper and all authors contributed to design of the experiments and data interpretation.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Prokhorova, I., Osterman, I., Burakovsky, D. et al. Modified nucleotides m2G966/m5C967 of Escherichia coli 16S rRNA are required for attenuation of tryptophan operon. Sci Rep 3, 3236 (2013). https://doi.org/10.1038/srep03236
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03236
This article is cited by
-
Structural and evolutionary insights into ribosomal RNA methylation
Nature Chemical Biology (2018)
-
Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly
Nature Structural & Molecular Biology (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.