Abstract
The Cyclin-dependent kinase 5 regulatory subunit-associated protein 1-like (CDKAL1) gene rs7756992 A/G polymorphism has been suggested to be associated with type 2 diabetes mellitus (T2DM), but the individual studies results are still controversial. To explore the association of CDKAL1 gene rs7756992 A/G polymorphism with T2DM, a meta-analysis involving 62,567 subjects from 21 separate studies was conducted. In the whole population, a significant association was found between CDKAL1 gene rs7756992 A/G polymorphism and T2DM under allelic (OR: 1.180, 95% CI: 1.130–1.230, P = 1.60 × 10−14), recessive (OR: 1.510, 95% CI: 1.380–1.660, P = 8.41 × 10−18), dominant (OR: 1.175, 95% CI: 1.109–1.246, P = 6.30 × 10−8), homozygous (OR: 1.400, 95% CI: 1.282–1.530, P = 8.02 × 10−14) and heterozygous genetic models (OR: 1.101, 95% CI: 1.040–1.166, P = 0.001). CDKAL1 gene rs7756992 A/G polymorphism was significantly associated with T2DM. The person with G allele of CDKAL1 gene rs7756992 A/G polymorphism might be predisposed to T2DM.
Similar content being viewed by others
Introduction
Type 2 diabetes mellitus (T2DM) is a chronic metabolic disturbance syndrome led by the combined actions of genetic gene and environmental factors. The data from International Diabetes Federation showed that in 2011, there were approximately 366 million diabetes mellitus (DM) patients in the globe and it has been speculated that the DM patients quantity will continually increase and be up to 552 million by 2030, of which the type 2 DM (T2DM) accounts for 90–95%1. The large scale investigations performed in the Chinese adults in June 2007 and May 2008 have shown that T2DM and impaired glucose tolerance (IGT) patients were about 92.4 million and 148 million respectively which are predominantly the young and middle-aged people and the number is in the first place of the world. Therefore, it is no time to delay to prevent T2DM2.
The interaction of genetic and environmental factors is universally acknowledged as the primary underlying T2DM mechanism. It is now generally considered that T2DM is not a sole disorder, but a multi-gene disorder with extensive heredity heterogeneity which results from the insulin resistance and β cell dysfunction of pancreatic island. The T2DM risk in the first degree relatives of T2DM patients is 3.62 times that in the common population3, so the researchers of various countries make great efforts to explore the T2DM susceptible genes. Once the T2DM susceptible genes are sought out, it means that the T2DM prevention clues have been found. It is an effective measure to screen the T2DM susceptible population and prevent T2DM progress.
It has been reported that more than 50 genes are closely associated with T2DM by genome wide association study (GWAS) technology4,5. Cyclin-dependent kinase 5 regulatory subunit-associated protein 1-like (CDKAL1) is one of the novel T2DM associated genes identified recently6. CDKAL1 gene, located in 6p22.3, spans 37 kb which encodes 579 amino acids. CDKAL1 gene encode tRNA decoration enzyme, namely methyl transfer enzyme which is responsible for the 2-methylthio-N6-threonylcarbamoyladenosine synthesis of the 37th base of tRNA Lys(UUU) 7. Wei et al found the mitochondria adenosine triphosphate (ATP) generation obstacle and the first stage insulin secretion impairment in the CDKAL1 gene knock-out mice8. In 2007, the GWAS study in Iceland population first reported that the CDKAL1 gene rs7756992 A/G polymorphism was associated with T2DM risk which was repeatedly verified in Caucasian populations6.
Although many studies on the relationship between CDKAL1 gene rs7756992 A/G polymorphism and T2DM have been performed so far, the researches results were still controversial. In 2011, Chistiakov et al found that the allele G of rs7756992 with higher diabetes risk thereby replicating the predisposing role of CDKAL1 gene in etiology of T2DM in a Russian population (OR = 1.21, 95% CI: 1.04–1.42, P = 0.017)9. In 2013, Li W et al found the similar result in a Chinese population (OR = 1.50, 95% CI: 1.11–2.04, P = 0.009)10. In contrast, in 2007, Horikoshi et al observed that in a Japan population with CDKAL1 gene rs7756992 GG genotypes, the T2DM risk was significantly decreased (OR:0.78, 95% CI: 0.61–0.98, P = 0.04)11. However, in 2010, Xu et al reported that CDKAL1 gene rs7756992 A/G polymorphism was not significantly associated with T2DM susceptibility in another Chinese population (OR: 1.68, 95% CI: 0.91–3.09, P = 0.10)12.
In the present study, a meta-analysis involving 26,120 T2DM patients and 36,447 controls from 21 separate studies was performed to estimate the relationship of CDKAL1 gene rs7756992 A/G polymorphism and T2DM.
Results
Studies and populations
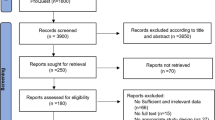
Twenty-eight publications were obtained through the retrieval process, among which fourteen manuscripts including twenty one studies met the inclusion criteria. Among the fourteen discharged papers, three papers were published repeatedly, four papers were of review character and five papers were not involved with CDKAL1 gene rs7756992 A/G polymorphism or T2DM. Two studies deviating from the Hardy-Weinberg equilibrium (HWE) were rejected. All of information was extracted from 26,120 T2DM cases and 36,447 controls (Table 1)6,9,10,11,12,13,14,15,16,17,18,19,20,21. One publication generally included one study. Additionally, some publications included multiple studies. For example, the manuscript published by Steinthorsdottir et al included five individual studies6. In addition, four individual studies were involved in the publication authored by Cauchi et al14. Thirteen countries were included in the present meta-analysis as China, Korea, Denmark, Iceland, Netherland, West Africa, Japan, France, Austria, United States, Morocco, Israel and Russia. These populations belong to Caucasian, Asian and African subgroups respectively. The Caucasian subgroup includes 7 individual studies, the Asian subgroup consists of 12 individual studies and the African subgroup comprises 2 individual studies.
Pooled analyses
In the whole population, a significant association was found between CDKAL1 gene rs7756992 A/G polymorphism and T2DM under allelic (OR: 1.180, 95% CI: 1.130–1.230, P = 1.60 × 10−14), recessive (OR: 1.510, 95% CI: 1.380–1.660, P = 8.41 × 10−18), dominant (OR: 1.175, 95% CI: 1.109–1.246, P = 6.30 × 10−8), homozygous (OR: 1.400, 95% CI: 1.282–1.530, P = 8.02 × 10−14) and heterozygous genetic models (OR: 1.101, 95% CI: 1.040–1.166, P = 0.001).
In the subgroup analysis, there was a significant association between them in Caucasian population under allelic (OR: 1.220, 95% CI: 1.170–1.270, P = 5.85 × 10−22), recessive (OR: 1.470, 95% CI: 1.340–1.610, P = 4.22 × 10−16), dominant (OR: 1.239, 95% CI: 1.176–1.305, P = 7.85 × 10−15), homozygous (OR: 1.530, 95% CI: 1.391–1.684, P = 5.81 × 10−18) and heterozygous genetic models (OR: 1.185, 95% CI: 1.122–1.252, P = 1.54 × 10−9).
In Asian subgroup analysis, a significant association between CDKAL1 gene rs7756992 A/G polymorphism and T2DM was also detected under allelic (OR: 1.190, 95% CI: 1.110–1.270, P = 3.58 × 10−7), recessive (OR: 1.570, 95% CI: 1.360–1.800, P = 3.85 × 10−10), dominant (OR: 1.173, 95% CI: 1.068–1.289, P = 8.68 × 10−4) and homozygous genetic models (OR: 1.404, 95% CI: 1.237–1.594, P = 1.52 × 10−7). No significant association was found under heterozygous genetic model (OR: 1.078, 95% CI: 0.990–1.174, P = 0.083).
In the African subgroup, there was no significant association between CDKAL1 gene rs7756992 A/G polymorphism and T2DM under allelic (OR: 1.070, 95% CI: 0.960–1.190, P = 0.23), dominant (OR: 1.008, 95% CI: 0.843–1.206, P = 0.927), homozygous (OR: 1.058, 95% CI: 0.840–1.332, P = 0.631), or heterozygous genetic models (OR: 0.954, 95% CI: 0.788–1.154, P = 0.626). Only under recessive genetic model, a significant association was found between them (OR: 1.420, 95% CI: 1.040–1.940, P = 0.03). (Table 2, Figure 1, 2, 3, 4, 5).
There was significant heterogeneity in the Asian subgroup under all of the genetic models (P < 0.05), while the heterogeneity did not exist under all of the genetic models in the Caucasian or African subgroup (P > 0.05). In order to explore the heterogeneity source, subsequent meta-regression was performed in the Asian population. Under the allelic, recessive and homozygous genetic models, the GG genotype number of T2DM group (GG1) was verified to be the main confounding factor to explain the heterogeneity source (P < 0.05).
According to the GG genotype of T2DM group, the Asian population was separated into two subgroups. The studies with GG1 > 200 were grouped to subgroup 1 and the residual studies with GG1 < 200 belonged to subgroup 2. In the following subgroup analysis stratified by GG1, significant increased T2DM risk was only detected in the subgroup 1 (allelic: OR: 1.240, 95% CI: 1.180–1.290, P = 2.85 × 10−22; recessive: OR: 1.670, 95% CI: 1.510–1.840, P = 4.52 × 10−24; homozygous: OR: 1.511, 95% CI: 1.407–1.624, P = 7.42 × 10−27). Under the recessive genetic model, a significant association was detected in subgroup 2 (OR: 1.470, 95% CI: 1.030–2.100, P = 0.03). Nevertheless, no significant association was detected in subgroup 2 under the allelic or homozygous genetic model (allelic: OR: 1.120, 95% CI: 0.970–1.300, P = 0.13; homozygous: OR: 1.272, 95% CI: 0.938–1.725, P = 0.121). Moreover, the heterogeneity was distinctly lower in subgroup 1 than that in the whole population (allelic: Pheterogeneity = 0.23, I2 = 27.4%; recessive: Pheterogeneity = 0.03, I2 = 58.7%; homozygous: Pheterogeneity = 0.234, I2 = 26.7%), while in subgroup 2, the heterogeneity still existed (allelic: Pheterogeneity = 0.0004, I2 = 78.0%; recessive: Pheterogeneity < 0.00001, I2 = 87.0%; homozygous: Pheterogeneity < 0.00001, I2 = 77.6%).
Under the dominant and heterozygous genetic models, AA genotype number of control group (AA0) was suggested to be the main heterogeneity source (P < 0.05). According to AA0, the Asian population was divided into two subgroups. Subgroup 1 was defined as AA0 > 300 and subgroup 2 was denoted as AA0 < 300. In the following subgroup analysis stratified by AA0, there was a significant association between CDKAL1 gene rs7756992 A/G polymorphism and T2DM in subgroup 1 (dominant: OR: 1.229, 95% CI: 1.123–1.346, P = 8.20 × 10−6; heterozygous: OR: 1.114, 95% CI: 1.022–1.215, P = 0.014), but no significant association between them was found in subgroup 2 (dominant: OR: 1.088, 95% CI: 0.886–1.335, P = 0.421; heterozygous: OR: 1.010, 95% CI: 0.839–1.217, P = 0.914). Although the heterogeneity still existed in subgroup 2 under the two genetic models (dominant: Pheterogeneity = 0.004, I2 = 70.7%; heterozygous: Pheterogeneity = 0.028, I2 = 60.2%), it was reduced and even did not exist any longer in subgroup 1 (dominant: Pheterogeneity = 0.033, I2 = 58.8%; heterozygous: Pheterogeneity = 0.086, I2 = 48.2%). (Table 3).
Bias diagnostics
The publication bias among the individual studies was evaluated by funnel plot and Egger's test. There was no visual publication bias in the Begg's funnel plot (Figure 6). There was no significant difference in the Egger's test yet, which suggested that no publication bias was detected in the current meta-analysis by using recessive genetic model (T = −0.29, P = 0.777). As no duplicate publications were included in the meta-analysis and every included individual study was a case-control study, there was no sample overlap in the cases or controls. In addition, as the controls data in each individual study were originally collected by the authors themselves and not cited from other studies, the samples in each of the studies were entirely independent and could not cause the results to be biased.
Discussion
In the current meta-analysis, a significant association was detected in the whole population between CDKAL1 gene rs7756992 A/G polymorphism and T2DM under allelic (OR: 1.18), recessive (OR: 1.51), dominant (OR: 1.175), homozygous (OR: 1.40) and heterozygous genetic models (OR: 1.101). In the subgroup analysis, there was a significant association in Caucasian and Asian subgroups (P < 0.05), while no significant association was detected in African subgroup (P > 0.05). In conclusion, it was indicated that the G allele of CDKAL1 gene rs7756992 A/G polymorphism might increase the T2DM risk, except in the African population. Moreover, the results in the whole population and Caucasian subgroup were genome-wide significant under most of the genetic models (P < 8.0 × 10−7). In Asian subgroup, the results reached the genome-wide significant threshold under allelic, recessive and homozygous genetic models. The negative results in the African population was perhaps not only associated with the different ethnicity, but also associated with the small sample size, because only two researches with 1350 T2DM subjects were included in this subgroup. In comparison to 9005 and 16,765 for the Caucasian and Asian studies respectively, the sample size1350 for the African studies was too small. Hence, the conclusion needs to be further verified by more and more studies with larger sample size in the African subgroup in the future.
In consideration of the significant heterogeneity in the Asian populations, the meta-regression has been performed. The confounding factor GG1 was confirmed to be the main heterogeneity source under the allelic, recessive and homozygous genetic models. AA0 was considered as the main heterogeneity source under the dominant and heterogeneity genetic models. In the subgroup analysis stratified by the two confounding factors under the five genetic models, the larger the confounding factors number (the subgroup in GG1 > 200, AA0 > 300), the smaller the heterogeneity (allelic: I2 = 27.4%; recessive: I2 = 58.7%; dominant: I2 = 58.8%; homozygous: I2 = 26.7%; and heterozygous: I2 = 48.2%), the stronger the association between CDKAL1 gene rs7756992 A/G polymorphism and T2DM risk (allelic: OR 1.240; recessive: OR 1.670; dominant: OR 1.229; homozygous: OR 1.511; heterozygous: OR 1.114). By contrast, the smaller the confounding factors number (the subgroup in GG1 < 200, AA0 < 300), the larger the heterogeneity (allelic: I2 = 78.0%; recessive: I2 = 86.9%; dominant: I2 = 70.7%; homozygous: I2 = 77.6%; and heterozygous: I2 = 60.2%), the weaker the association between them (allelic: OR 1.120; recessive: OR 1.470; dominant: OR 1.088; homozygous: OR 1.272; heterozygous: OR 1.010). It suggested that the larger sample size could reduce the heterogeneity between the individual studies and the research sample size needs to be further expanded in the future.
Cyclin-dependent kinase 5 (CDK5) is a serine/threonine protein kinase. The CDK5 is activated by producing CDK5/p35 compounds in the pancreatic tissue, thus the β cells are degenerated and the insulin secretion is inhibited, especially in the high glucose condition22. In 2006, Ubeda et al found that in the high glucose internal environment, the CDK5 overactivity could decrease the insulin release rate and reduce the insulin production and restrain the insulin gene expression. They found that inhibition of CDK5 activity could protect pancreatic cells from glucotoxity23.
CDKAL1 is highly expressed in the human pancreas, skeletal muscle and brain tissue and can specially inhibit CDK5 activity24. In 2010, Ohara-Imaizumi et al found that CDKAL1 controls first-phase insulin exocytosis in β cells by facilitating ATP generation, K (ATP) channel responsiveness and the subsequent activity of Ca (2+) channels through pathways other than CDK5-mediated regulation.CDKAL1 gene rs7756992 A/G mutation probably leads to the inhibition effect loss on CDK5, thus the T2DM risk is increased25. However, the exact underlying mechanism of CDKAL1 gene mutation changing the insulin secretion pattern are still unclear and need to be clarified in the further researches.
No similar meta-analysis on the association of T2DM with CDKAL1 gene rs7756992 A/G polymorphism has been found internationally so far. Some limitations still existed in the present meta-analysis. Large-scale researches on the association of T2DM with CDKAL1 gene rs7756992 A/G polymorphism are still inadequate. The serum CDKAL1 level was influenced not only by the CDKAL1 gene rs7756992 A/G polymorphism, but also by other gene polymorphism as rs7754840 G/C polymorphism and unscientific dietary habits20. As the heredity model of T2DM is multiple gene inheritance which means that many micro-effect genes produce a general effect and leads to T2DM, other genes polymorphisms might be predisposed to T2DM risk26.
Finally, CDKAL1 gene rs7756992 A/G polymorphism was significantly associated with T2DM susceptibility, particularly in the Caucasian and Asian population. The persons with the G allele of CDKAL1 gene rs7756992 A/G polymorphism might be predisposed to T2DM. This conclusion might guide us to formulate new T2DM therapy strategy. Taken account the above limitations, more studies on the association of CDKAL1 gene rs7756992 A/G polymorphism and T2DM are needed to be carried out to further clarify the conclusion in the future.
Methods
Publication search and inclusion criteria
The electronic databases including PubMed, Web of Science, Embase, China Biological Medicine Database and China National Knowledge Infrastructure were searched by using the words as “CDKAL1”, “rs7756992”, “polymorphism” and “type 2 diabetes mellitus”. The last research was updated on October 15, 2013 with the publication years ranging from 2007 to 2013.
The following major criteria should be met by the included studies. The CDKAL1 gene rs7756992 A/G polymorphism and T2DM.were evaluated. T2DM was diagnosed by the American Diabetes Association fasting plasma criteria (2005). The fasting plasma glucose level was no less than 7.0 mmol/L or the 2 h plasma glucose of oral glucose tolerance test was no less than 11.1 mmol/L. Furthermore, no genetic relationship existed between the subjects in the individual studies. Only the data drawn from officially published manuscripts with case-control or cohort studies could be adopted. The HWE should be followed by genotype member of the control group in the individual studies.
Data extraction
The informed consent was obtained from all subjects. The studies data was extracted in the light of a standard protocol. Three researchers carried out the meta-analysis; two of whom searched out the studies duplicately and the third researcher acted as the arbitrator to resolve the conflict between the two researchers and come to an agreement. The current meta-analysis rejected the studies that did not follow the selection criteria, that were repeatedly published, or that provided insufficient data. If similar data was rooted in different manuscripts by the same authorship, the data was only once adopted. The following items including the first author's name, publication year, region, continents, number of genotypes, matching criteria and total number of cases and controls should be shown in the extracted data.
Statistical analyses
Five genetic models as the allelic (G allele distribution frequency), recessive (GG vs. AA + AG), dominant (GG + AG vs. AA), homozygous (GG vs. AA) and heterozygous (AG vs. AA) genetic models were used in the present meta-analysis. The odds ratios (ORs) and their corresponding 95% confidence intervals (CIs) were used to compare the association of CDKAL1 gene rs7756992 A/G polymorphism and T2DM. The heterogeneity among the studies was calculated by Chi-square-based Q-tests with significance set at P < 0.05 level27. If heterogeneity existed, the random-effect model (DerSimonian and Laird method) would be used28. Or else, the fixed-effect model was adopted (the Mantel–Haenszel method)29. Z test was used to estimate the pooled OR with significance set at P < 0.05 level.
The HWE was assessed by using Fisher's exact test with significance set at P < 0.05 level. The potential publication bias was estimated by adopting the funnel plot. The funnel plot symmetry was evaluated by using Egger's linear regression test on the natural logarithm scale of the OR and significance was set at P < 0.05 level30. The statistical analyses were performed by Stata 12.0 software (StataCorp, College Station, TX, USA).
References
Whiting, D. R., Guariguata, L., Weil, C. & Shaw, J. IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract 94, 311–21 (2011).
Yang, W. et al. Prevalence of diabetes among men and women in China. N Engl J Med 362, 1090–101 (2010).
Shen, H. B. et al. An epidemiological study on genetic agent in type 2 diabetes mellitus. China Public Health 15, 492–4 (1999).
Scott, L. J. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316, 1341–5 (2007).
Li, H. et al. A genome-wide association study identifies GRK5 and RASGRP1 as type 2 diabetes loci in Chinese Hans. Diabetes 62, 291–8 (2013).
Steinthorsdottir, V. et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 39, 770–5 (2007).
Pierrel, F., Douki, T., Fontecave, M. & Atta, M. MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J Biol Chem 279, 47555–63 (2004).
Wei, F. Y. et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest 121, 3598–608 (2011).
Chistiakov, D. A. et al. The carriage of risk variants of CDKAL1 impairs beta-cell function in both diabetic and non-diabetic patients and reduces response to non-sulfonylurea and sulfonylurea agonists of the pancreatic KATP channel. Acta Diabetol 48, 227–35 (2011).
Li, W. et al. Association analysis of CDKAL1 rs7756992 polymorphism with the clinical characteristics and susceptibility. Journal of Wenzhou Medical College 43, 141–6 (2013).
Horikoshi, M. et al. Variations in the HHEX gene are associated with increased risk of type 2 diabetes in the Japanese population. Diabetologia 50, 2461–6 (2007).
Xu, M. et al. Combined effects of 19 common variations on type 2 diabetes in Chinese: results from two community-based studies. PLoS One 5, e14022 (2010).
Omori, S. et al. Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8 and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes 57, 791–5 (2008).
Cauchi, S. et al. Post genome-wide association studies of novel genes associated with type 2 diabetes show gene-gene interaction and high predictive value. PLoS One 3, e2031 (2008).
Liu, Y. et al. Positive association between variations in CDKAL1 and type 2 diabetes in Han Chinese individuals. Diabetologia 51, 2134–7 (2008).
Ng, M. C. et al. Implication of genetic variants near TCF7L2, SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2 and FTO in type 2 diabetes and obesity in 6,719 Asians. Diabetes 57, 2226–33 (2008).
Horikawa, Y. et al. Replication of genome-wide association studies of type 2 diabetes susceptibility in Japan. J Clin Endocrinol Metab 93, 3136–41 (2008).
Rong, R. et al. Association analysis of variation in/near FTO, CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761 and CDKN2B with type 2 diabetes and related quantitative traits in Pima Indians. Diabetes 58, 478–88 (2009).
Takeuchi, F. et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes 58, 1690–9 (2009).
Tabara, Y. et al. Replication study of candidate genes associated with type 2 diabetes based on genome-wide screening. Diabetes 58, 493–8 (2009).
Lu, F. et al. Genetic variants on chromosome 6p21.1 and 6p22.3 are associated with type 2 diabetes risk: a case-control study in Han Chinese. J Hum Genet 57, 320–5 (2012).
Ching, Y. P., Leong, V. Y., Wong, C. M. & Kung, H. F. Identification of an autoinhibitory domain of p21-activated protein kinase 5. J Biol Chem 278, 33621–4 (2003).
Ubeda, M., Rukstalis, J. M. & Habener, J. F. Inhibition of cyclin-dependent kinase 5 activity protects pancreatic beta cells from glucotoxicity. J Biol Chem 281, 28858–64 (2006).
Wei, F. Y. et al. Cdk5-dependent regulation of glucose-stimulated insulin secretion. Nat Med 11, 1104–8 (2005).
Ohara-Imaizumi, M. et al. Deletion of CDKAL1 affects mitochondrial ATP generation and first-phase insulin exocytosis. PLoS One 5, e15553 (2010).
Li, Y. Y. ENPP1 K121Q polymorphism and type 2 diabetes mellitus in the Chinese population: a meta-analysis including 11,855 subjects. Metabolism 61, 625–33 (2012).
Cochran, W. G. The effectiveness of adjustment by subclassification in removing bias in observational studies. Biometrics 24, 295–313 (1968).
DerSimonian, R. & Laird, N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–88 (1986).
Mantel, N. & Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–48 (1959).
Egger, M., Davey, G., Schneider, M. & Minder, C. Bias in meta-analysis detected by a simple, graphical test. British Medical Journal 315, 629–34 (1997).
Acknowledgements
This work was funded by the National Natural Science Foundation of China (NSFC 81100073 to Dr Yan-yan Li), Excellent Young and Middle-Aged Teachers Assistance Program of Nanjing Medical University for Dr Yan-yan Li (2013-2015, JX2161015034) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). Thank all our colleagues working in the Department of geriatrics, the First Affiliated Hospital of Nanjing Medical University.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: Y.L. Performed the experiments: Y.L., L.W. Analyzed the data: Y.L., C.Z., Z.Y. Contributed reagents/material/analysis tools: Y.L., J.X. Wrote the manuscript: Y.L., Y.Q. Reference collection and data management: Y.L., X.W. Statistical analyses and paper writing: Y.L., X.L. Study design: Y.L., A.C.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Li, Yy., Wang, Ls., Lu, Xz. et al. CDKAL1 gene rs7756992 A/G polymorphism and type 2 diabetes mellitus: a meta-analysis of 62,567 subjects. Sci Rep 3, 3131 (2013). https://doi.org/10.1038/srep03131
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03131
This article is cited by
-
A critical review on therapeutic approaches of CRISPR-Cas9 in diabetes mellitus
Naunyn-Schmiedeberg's Archives of Pharmacology (2023)
-
Body mass index modulates the association between CDKAL1 rs10946398 variant and type 2 diabetes among Taiwanese women
Scientific Reports (2018)
-
Association of CDKAL1 nucleotide variants with the risk of non-syndromic cleft lip with or without cleft palate
Journal of Human Genetics (2018)
-
A54T polymorphism in the fatty acid binding protein 2 studies in a Saudi population with type 2 diabetes mellitus
Lipids in Health and Disease (2014)
-
The association between the rs6495309 polymorphism in CHRNA3 gene and lung cancer risk in Chinese: a meta-analysis
Scientific Reports (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.