Abstract
Breast cancer is the most frequent malignancy affecting women worldwide. It has been suggested that infection by Epstein Barr Virus (EBV), Mouse Mammary Tumor Virus or a similar virus, MMTV-like virus (MMTV-LV), play a role in the etiology of the disease. However, studies looking at the presence of these viruses in breast cancer have produced conflicting results and this possible association remains controversial. Here, we used polymerase chain reaction assay to screen specific sequences of EBV and MMTV-LV in 86 tumor and 65 adjacent tissues from Mexican women with breast cancer. Neither tumor samples nor adjacent tissue were positive for either virus in a first round PCR and only 4 tumor samples were EBV positive by a more sensitive nested PCR. Considering the study's statistical power, these results do not support the involvement of EBV and MMTV-LV in the etiology of breast cancer.
Similar content being viewed by others
Introduction
Breast cancer (BC) is one of the most frequent malignancies affecting women worldwide, with an estimated 1.38 million new cases diagnosed in 2008, representing 23% of all cancers1. Risk factors include a family and personal history of BC, lifetime menstrual cycles, reproductive history, hormone therapy, cigarette smoking, obesity and others2. Viral infection has also been proposed to trigger the development of BC3,4,5,6. Epstein Barr virus (EBV) has been consistently associated with several malignancies, including Burkitt's and Hodgkin's lymphoma, nasopharyngeal carcinoma (NPC) and some studies also support a link with BC. EBV initially raised interest as a possible causal agent in BC because the high incidence of male BC observed in countries with a high frequency of EBV-associated lymphomas4,7 and because of the similar histological pattern between NPC and medullary BC8. The first evidence of the possible participation of EBV in BC was reported by Labreque et al, who found EBV sequences in 21% of BC samples7. However, other studies have generated contradictory results, with association frequencies ranging from 0 to 100%7,9,10,11,12,13, probably reflecting the lack of uniformity in the study design and methods by the different authors (for a comprehensive analysis of the methodology utilized see14).
The non-acute transforming beta retrovirus mouse mammary tumor virus (MMTV) is responsible for most BC cases in several strains of mice15. This observation prompted the search for MMTV-like sequences (MMTV-LS) in human BC, under the assumption that MMTV or a related virus (MMTV-LV) could be involved. Reports addressing the presence of MMTV-LS in human BC have also generated highly variable results, with some studies reporting frequencies of up to 80–100%3,5,16,17,18 and others unable to find evidence of infection1,6,19,20.
In this study, we screened for both EBV and MMTV-LV sequences in BC tissue samples from Mexican patients by a standard polymerase chain reaction (PCR) assay. Our viral screening tests are based on what is known about the mechanisms of EBV and MMTV oncogenesis, which are through direct infection of the cancer initiating cell. Thus, if infection is part of the initial genetic lesion and cancer is a monoclonal expansion of this initiating cell, then all tumor cells should carry evidence of the viral infection: EBV episomes or MMTV-LS integrated in the cellular genome. In order to make the test more stringent we included in the analysis only samples with ≥30% of tumor cells and designed a PCR test to equate the number of infected cells with the number of tumor cells. Also, since some retroviruses show high sequence variability, we have designed two separated PCRs to interrogate for the presence of MMTV-LS. Furthermore, we have compared tumor samples with non-tumor control samples isolated from the same patients and a more sensitive nested PCR was used to confirm results from both EBV and MMTV-LS tests. With this strategy we have found no evidence of participation of either EBV or an MMTV-LV in Mexican samples of BC.
Results
Study population
A description of the patients included in this study is presented in Table 1. Sixty-five cases were recruited in the Oncology and Gynecology hospitals in Mexico City and 21 in the General hospital in Merida, for a total of 86 cases. Non-tumor adjacent-tissue was obtained in all 65 cases from Mexico City hospitals, but not in patients from Merida. Median age for patients in Mexico City was 60 years and 50 years for patients of Merida. Most cases presented with infiltrating ductal carcinoma (67.4%) and less with mixed carcinoma (16.3%), in situ ductal carcinoma (8.1%), or infiltrating lobular carcinoma (8.1%). Only tissues of ≥30 tumor cells were included in the study; tissues ranged from 30 to 95% tumor cells with a mean = 56%. Figure 1 shows two examples of tissues with about 55–60% of tumor cells.
EBV screening
The limit of detection of the PCR for EBV was about 900 infected cells in the first PCR (Figure 2a) in Raji cells, which have been reported to carry about 50 EBV episomal copies per cell. A similar number has been reported for EBV-associated lymphomas21. This would mean that our PCR test is able to detect down to 45,000 viral genomes. Similar detection limits were obtained using Daudi and B95-8 cell lines that carry slightly different number of EBV episomes (not shown). The number of EBV copies per cell in EBV-associated carcinomas is not known, although a study in a single patient with NPC estimated 7 copies/cell22. 200 ng of DNA (equivalent to about 30,000 cells) were analyzed in the first PCR. Since samples had ≥30% tumor cells, we were testing ≥9,000 tumor cells per sample. Assuming that breast carcinoma cells also harbor 7 EBV copies/cell, our sample would contain ≥63,000 viral genomes, which is in the order of the limit of detection of the assay of 45,000 viral genomes if infection is part of the initiating oncogenic insult and therefore all or most tumor cells are infected. Although the number of EBV copies/cell in breast epithelial cells is just an educated guess, the first PCR test was able to detect EBV genomes from a gastric cancer lymphoepitheliome type (Fig. 2c) and we have used this PCR to screen GC tissues, finding about 10% of EBV positives (submitted for publication). Furthermore, we confirmed the first PCR with a more sensitive nested PCR, which detects EBV in ≥30 Raji cellular genomes (Fig. 2b). Still, we considered that only samples positive in the first PCR would support a viral participation in the tumor genesis.
Limit of detection for the EBV PCRs.
(a) Limit of detection of first PCR expressed in number of cellular genomes (Raji cells) and EBV genomes. White arrows indicate the lower limits of detection set in 900 cells for the first PCR and, (b) of 30 cells for the nested PCR. (c) Detection of EBV genome in a sample of gastric cancer type lymphoepitheliome (LE). DNA from EBV positive cell line Raji was used as positive control (C+).DNA from the EBV negative cell line Ramos was used as negative control (C−). Molecular marker (M).
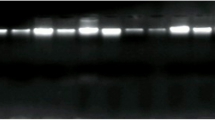
Once the limit of detection was established, we analyzed each tumor and non-tumor tissue by PCR and nested PCR. We did not observe any positive sample after the first PCR (Fig. 3a), whereas we found four (4.7%) positive samples with the nested PCR (Fig. 3b). Because only the first PCR equates the number of tumor cells, these data do not support an EBV participation in the tumorigenic process. None of the adjacent non-tumor tissues were EBV positive after the first and nested PCRs. The four products by nested PCR were purified and sequenced and their EBV identity was confirmed.
Screening of EBV in breast cancer samples.
(a) Ten representative samples of BC analyzed by the first PCR are shown and (c) by the nested PCR, including the four samples that gave a positive signal in the nested PCR. The β-actin cellular gene is shown as a control for DNA integrity (b). DNA from Raji cell line was used as positive control (C+) and Ramos as negative control (C−). Molecular marker (M).
MMTV screening
Considering the high rate of mutation of some retrovirus, for MMTV-LS screening, we implemented the following strategy to reduce false negatives resulting from inadequate primer recognition. First, the PCR detection was set up using two DNA templates, plasmid DNA (pENV) and genomic DNA from BALB/c mice spleen cells (mSP). The latter was included to more closely reproduce the high sequence variability found in vertically transmitted MMTV. Second, we carried out two first PCR tests targeting two different sequences in the MMTV env gene. Third, in order to favor primers hybridization even in the presence of mismatches, the annealing temperature was 50°C for both reactions; the above because temperature gradients showed that a good PCR signal could be obtained in a range of annealing temperatures from 48 to 58°C for primers P1–P4 (Fig. 4a) and from 47–57°C for primers P2–P3. The limits of detection of the first and nested PCRs for MMTV-LS were established as described for EBV. The limit of detection of the first PCRs was about 2100 mSP cells for both P1–P4 and P2–P3 reactions (not shown) whereas for pENV it was 25 plasmid copies for P1–P4 (Fig. 4b) and 250 for P2–P3 (Fig. 4c). Since MMTV genomes in lymphoid cells of virtually all laboratory mouse strains has been estimated between 2 to 8 copies per cell15, the limit of provirus detection is between 4,200–16,800. There are many more copies of the MMTV provirus in mouse BC samples, which has been estimated between 10–10023. We analyzed DNA equivalent to 9,000 (30% = 2700) tumor cells, which would be sufficient to detect MMTV infection even if as few as 2 (5,400) or 10 (27,000) provirus were harbored per tumor cell. In addition, a nested PCR that had an increased detection of at least 1000-fold was used to confirm the first PCR result.
MMTV screening PCRs.
(a) A gradient of annealing temperatures for the P1–P4 reaction is shown, with a detectable signal observed in the range of 48 to 58°C; a 50°C temperature (white arrow) was used to allow template recognition even in the presence of mismatches. (b) and (c) show the detection limits of the two first PCRs expressed in plasmid copy numbers; a signal can be seen in the range of 25 copies in panel (b) (product of 668 bp using primers P1–P4) and 250 copies in panel (c) (product of 253 bp using primers P1–P3). A PCR reaction without DNA was included as negative control (C−). Residual primers from the reaction are indicated with an arrowhead.
The 86 BC samples were analyzed with the two first round PCRs and no evidence of MMTV-LS infection was found in either, the tumor or non-tumor adjacent tissues. Figure 5a shows results of a screening of 10 BC samples with P1–P4 primers and Figure 5b with P2–P3 primers. Furthermore, none of the BC or non-tumor adjacent tissues were positive in the more sensitive nested PCR; Figure 5d shows results of the analysis of six representative BC samples.
Screening of MMTV in breast cancer samples.
First round PCR for ten representative samples of BC with P1–P4 primers (a) and P2–P3 primers (b). (c) PCR from endogenous β-actin gen. (d) Nested PCR of six BC samples. DNA from mouse spleen (mSP) and from plasmid (pENV) were used as positive controls and a PCR reaction mix without DNA was used as negative control (C−). In figure 5a an asterisk denotes an unspecific band.
Discussion
The involvement of different viruses in the etiology of human BC has been extensively investigated with highly variable results, providing evidence in favor or against it, hence the issue remains controversial. Different arguments have been used to explain the source of variability, including the high retroviral mutation rate accounting for false negatives to DNA contamination or the use of highly sensitive detection techniques accounting for false positives14. In this study, we analyzed 86 cases of breast cancer from Mexican patients and found evidence of EBV in 4 samples but no evidence of MMTV-LS in any of the specimens; still, the low level EBV signal found does not support clonal expansion of an altered cell in which viral infection played a role in oncogenesis. The screening approach used in this study was designed to test a number of tumor cells exceeding the sensitivity of the first PCR, since we would expect all tumor cells to be infected. The above because the documented oncogenic mechanisms for both MMTV and EBV are via direct infection, in the case of MMTV through insertional mutagenesis24 and expression of the viral env oncogene25,26 and for EBV mainly by expression of LMP1 and LMP2A viral oncogenes27,28,29. In agreement, most murine breast cancer tissues carry MMTV sequences in all tumor cells and in EBV-associated lymphomas and carcinomas, all tumor cells carry viral episomes. Furthermore, EBV positive-lymphoma cell lines fail to maintain the transformed state after experimental elimination of the EBV genome28. These data support that in MMTV and EBV associated neoplasias, cancer results from the clonal expansion of a wounded cell in which infection was part of the oncogenic damage. Further elimination of the virus is rather difficult since MMTV gets integrated in the host genome and EBV expresses EBNA1, a protein coordinating equally segregation of viral episomes to daughter cells. Highly sensitive techniques such as nested PCR, real-time PCR or Southern blot, often do not match infection with the total number of tumor cells in the sample. Here, we included in the analysis only samples with ≥30% of tumor cells and designed a PCR test to equate the number of infected cells with the number of tumor cells. A similar approach used in gastric cancer cell lines has shown 10% of EBV positive samples (submitted for publication). Also, since retroviral sequences show high variability, we have designed two separated PCRs to interrogate for the presence of MMTV-LS and the annealing temperature of the screening PCR was set up to allow for template mismatches. With this strategy we aimed to reduce the chances of both false positives and negatives.
Indirect mechanisms of transformation triggered by other pathogens have been described, such as chronic inflammation by Helicobacter pylori30,31 or immunosuppression by HIV32, in which the pathogen does not reside in the tumor cell. In the case of EBV or MMTV-LV participation in human BC, the hypothesis of an indirect transforming mechanism has not been documented with the appropriate experiments. The above are important considerations in the test design and in the interpretation of results when searching for possible virus-cancer associations. In our hands, no MMTV positive samples were found even with the more sensitive nested PCR; while 4.7% of the tumor samples were positive for EBV. However, as discussed above, we did not consider those cases as causally associated. It is well documented that about 95% of the adult world population is infected by EBV, usually residing in latently infected memory B cells2. Thus, we think that in those positive samples by the highly sensitive test, the signal does not originate in the tumor cells and most likely comes from infected B cells infiltrating the tumor lesion. 1–10 per million EBV infected blood cells have been estimated in healthy individuals33. Alternative explanations for tumor samples with low infection levels are: infected B cells in which the lytic cycle was triggered feeding the tumor with viral particles, as it has been previously documented34, or increased tropism of EBV for certain types of tumor cells, as suggested by quantitative PCR reports showing that breast cancer samples positive for EBV have very low levels of infection35,36.
The involvement of another murine retrovirus, the xenotropic murine leukemia-related virus or XMRV, in human disease highlighted how common is to contaminate tissues in the laboratory. XMRV was originally cloned from prostate cancer cell lines and several studies linked the virus with this cancer and chronic fatigue syndrome3,5,18,37. However, it was found later that XMRV originated as a result of genetic recombination between human and murine endogenous retroviruses after human cells were implanted in mice17. Currently, it is widely accepted that all XMRV positive associations resulted from sample contamination.
Finally, it is unlikely that our results were due to an insufficient sample size since we calculated the statistical power of the study and found that in our population (N = 86), the probability of finding at least one EBV or MMTV-LS positive sample starting from hypothetical frequencies of 10% and 5% was as high as 99.99% and 95.00% respectively. In conclusion, given the strict methodological approach and the high statistical power, our study does not support the participation of neither EBV nor MMTV-LV in Mexican samples of human breast cancer. Importantly, in all the tissues tested we did not have available any medullary type of cancer. This type of cancer resembles histologically the LE carcinomas, in which EBV has been associated with an 80–100% frequency. It is possible that EBV is mainly associated with this particular type of BC.
Methods
Ethics statement
This study was approved by the ethical and scientific review boards of the Instituto Mexicano del Seguro Social (IMSS): Comisión Nacional de Investigación Científica and Comité de Ética en Investigación. Norma Oficial Mexicana NOM-062-ZOO-1999, approved by COFEPRIS (register number CONBIOETICA09CEI01520130424). All patients prospectively enrolled were informed on the nature of the study and those willing to participate signed a written informed consent prior to specimen collection and were treated according to the ethical guidelines of our institution. The use of BALB/c mice was also approved by the ethical and scientific review boards of the Instituto Mexicano del Seguro Social (IMSS); animals were sacrificed by cervical dislocation before spleen isolation in order to ameliorate suffering in accordance with approved guidelines established by IMSS and the NIH Guide for the Care and Use of Laboratory Animals.
Study population
Breast cancer patients were recruited from the Oncology Hospital (Centro Medico Nacional Siglo XXI, CMN-SXXI) and the Gynecology Hospital “Luis Castelazo Ayala” in Mexico City and the General Hospital in Merida Yucatan in the South East of Mexico, all hospitals from IMSS (Table 1). In the Oncology and Gynecology Hospital all patients were enrolled prospectively, while samples from the General Hospital were from retrospective cases.
Histopathological examination
A tissue sample from the tumor was obtained from each patient and in patients from Oncology and Gynecology Hospital an additional sample was obtained from tissue adjacent to the tumor (at least 2 cm apart from the tumor lesion). Tissue samples were obtained in the surgical room and kept frozen at −70°C until used. In the cases from the Oncology and Gynecology hospitals, the frozen tissues were used for DNA extraction, whereas in the cases from the General Hospital, paraffin embedded tissues were used (Table 1). A fragment of all tissues was fixed in formaldehyde, embedded in paraffin and a slide preparation stained with hematoxylin-eosin (H-E) to determine the type and grade of the lesion and the frequency of tumor cells. The latter was done by the experienced pathologist of the Oncology Hospital (CMN SXXI IMSS). Only tissues containing ≥30 tumor cells were included in the study.
Cells, mice and plasmids
The cell lines utilized were: three EBV positive -Raji, Daudi and B95-8- and two EBV negative -Ramos and BJAB-; BJAB cell line was purchased from Life Technologies, Inc and all the others from the American Type Culture Collection. All cell lines were cultured in advanced RPMI medium supplemented with 4% of fetal bovine serum and Hepes 1× (all from Gibco, Carlsbad, CA) and maintained at 37°C in 5% of CO2. Spleens from BALB/c mice (kindly provided by Dr. K. Chávez, Pediatric Hospital at the CMN SXXI, IMSS) were removed in aseptic conditions and splenocytes were recovered by passing through a sterile mesh and lysing erythrocytes with lysing buffer (Sigma Aldrich, St. Louis Missouri, USA). To construct the env plasmid (pENV), a 687 bp env fragment was PCR amplified from BALB/c splenocytes using primers 1 and 4 previously described by Wang et al3 and cloned into the T-easy pGEM plasmid (Promega, Madison, USA) according to manufacturer's instructions.
DNA purification
In the case of cell lines and mice splenocytes, DNA was extracted from 5 × 106 cells using the QIAamp mini kit, whereas for frozen tissue, up to 10 mg were disrupted in the TissueLyser II for 20 seconds and DNA extracted with QIAamp DNA mini kit. For the paraffin embedded tissue samples, sections of 5–10 μm thick were cut and DNA purified using the QIAmp DNA FFPE tissue kit. Plasmid DNA was purified using QIAGEN Plasmid Mini Kit. All DNA purification kits were from QIAGEN (Hilden, Germany). Purified DNA was quantified using a spectrophotometer NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA), whereas DNA quality was determined by the 260/280 ratio and Integrity by PCR of β-actin and β-globin endogenous genes.
PCRs of endogenous genes
The PCR primers for β-actin were, forward 5′ CCT AAG GCC AAC CGT GAA AAG 3′ and reverse 5′ TCT TCA TGG TGC TAG GAG CCA 3′20 and for β-globin were, forward 5′ ACA CAA CTG TGT TCA CTA GC 3′ and reverse 5′ TGG TCT CCT TAA ACC TGT CTT G 3′. For both genes, the reaction mixture contained 100 ng of DNA in PCR buffer, 1.5 mM MgCl2, 200 μM dNTPs, 200 nM of each primer and 2.5 U of Taq Polymerase (all PCR reagents were from Thermo Fisher Scientific, Waltham, MA). For β-actin the amplification conditions were: an initial denaturation step of 7 min at 94°C, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 1 min, with a final extension step of 72°C for 10 min. For β-globin, the amplification conditions were: an initial denaturation step of 3 min at 95°C and then 30 cycles of 95°C for 45 sec, 54°C for 1 min and 72°C for 30 sec, followed by a final extension at 72°C for 2 min. PCR products were analyzed by electrophoresis in agarose gels.
Viral detection by standard and nested PCR
For the EBV PCR tests DNA from the infected cell lines Raji, Daudi and B95-8 was used as positive control and DNA from the uninfected BJAB and Ramos cells as negative control. For the standard first PCR primers LLW1 y LLW2 were used, which amplify a fragment (position 505 to 741 nucleotide) of the EBV BamHI W fragment7. This fragment is repeated 11 times in the EBV genome, which increases the sensitivity of EBV detection. The reaction mixture contained 200 ng of DNA in PCR buffer, 2.5 mM MgCl2, 200 μM dNTPs, 200 nM of each primer and 2.5 U of Taq Polymerase. The PCR reaction was: an initial denaturation step of 5 min at 94°C and then 30 cycles of 94°C for 1.5 min, 57°C for 45 sec and 72°C for 1 min, followed by a final extension of 72°C for 7 min (all PCR reagents were from Thermo Fisher Scientific, Waltham, MA). A nested PCR was designed with first PCR amplicon internal primers LLW1 int 5′ CTT TGT CCA GAT GTC AGG GG 3′ and LLW2 int 5′ GCC TGA GCC TCT ACT TTT GG 3′. PCR reactions contained 0.05 μl of the first PCR product (1:1000 dilution) and PCR buffer, 2.5 mM MgCl2, 200 μM dNTPs, 400 nM of each primer and 2.5 U of Taq Polymerase. The nested reaction was performed with an initial denaturation step at 94°C for 5 min, followed by 15 cycles consisting of 94°C for 20 sec, 57°C for 20 sec and 72°C for 30 sec and a final extension of 72°C for 7 min.
For MMTV screening we selected four primers (P1, P2, P3 and P4) previously described by Wang et al3, which target the MMTV env gene and are characterized by having low identity to human endogenous retroviral sequences (HERVs)3. According to MMTV NCBI Reference Sequence (NC_001503.1), primers P1 and P4 generate a fragment of 668 bp and primers P2 and P3 generate a fragment of 253 pb, however, the expected amplicon size may slightly vary due to quasispecies variation. The first PCR reactions were performed with P1–P4 and P2–P3 primers, whereas primers P2–P3 were used for the nested PCR to amplify an internal fragment of the first P1–P4 PCR product. PCR reaction components were as described for the endogenous genes, except for the amount of DNA. In the case of plasmid DNA we used 2.9 × 10−4 ng and for mouse DNA 50 ng; whereas for the nested PCR we used 0.05 μl of first P1–P4 PCR product (1:1000 dilution). The cycling conditions were: an initial denaturation step of 5 min at 94°C; 35 (first round) or 20 (nested) cycles consisting of 94°C for 45 sec, 50°C for 1.5 min and 72°C for 45 sec, followed by a final extension at 72°C for 10 min.
Sequencing
Positive samples were confirmed by sequencing of both forward and reverse strands of the PCR products, after purification with QIAquick gel extraction kit (Qiagen Hilden, Germany). Sequences were compared with the Genebank database38 using the BLAST program39. Sequencing was carried out in the Instituto de Biología, Universidad Nacional Autónoma de México.
Statistical analysis
The statistical power of the findings was evaluated in relation to the ability of a study of this sample size (N = 86) to detect viral genomes (infecting the bulk of the tumor cells) in at least one patient. The hypothetical frequencies of infection were 10% and 5%. The statistical package used for this analysis was Epi Info version 7.1.240.
References
Bindra, A. et al. Search for DNA of exogenous mouse mammary tumor virus-related virus in human breast cancer samples. The Journal of general virology 88, 1806–1809, 10.1099/vir.0.82767-0 (2007).
Dumitrescu, R. G. & Cotarla, I. Understanding breast cancer risk -- where do we stand in 2005? Journal of cellular and molecular medicine 9, 208–221 (2005).
Wang, Y. et al. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer research 55, 5173–5179 (1995).
Sasco, A. J., Lowenfels, A. B. & Pasker-de Jong, P. Review article: epidemiology of male breast cancer. A meta-analysis of published case-control studies and discussion of selected aetiological factors. International journal of cancer. Journal international du cancer 53, 538–549 (1993).
Zapata-Benavides, P. et al. Mouse mammary tumor virus-like gene sequences in breast cancer samples of Mexican women. Intervirology 50, 402–407, 10.1159/000110652 (2007).
Park, D. J., Southey, M. C., Giles, G. G. & Hopper, J. L. No evidence of MMTV-like env sequences in specimens from the Australian Breast Cancer Family Study. Breast cancer research and treatment 125, 229–235, 10.1007/s10549-010-0946-4 (2011).
Labrecque, L. G., Barnes, D. M., Fentiman, I. S. & Griffin, B. E. Epstein-Barr virus in epithelial cell tumors: a breast cancer study. Cancer research 55, 39–45 (1995).
Lespagnard, L. et al. Absence of Epstein-Barr virus in medullary carcinoma of the breast as demonstrated by immunophenotyping, in situ hybridization and polymerase chain reaction. American journal of clinical pathology 103, 449–452 (1995).
Gaffey, M. J. et al. Medullary carcinoma of the breast. Identification of lymphocyte subpopulations and their significance. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc 6, 721–728 (1993).
Chu, J. S., Chen, C. C. & Chang, K. J. In situ detection of Epstein-Barr virus in breast cancer. Cancer letters 124, 53–57 (1998).
Preciado, M. V. et al. Epstein-Barr virus in breast carcinoma in Argentina. Archives of pathology & laboratory medicine 129, 377–381, 10.1043/1543-2165(2005)129377:evibci>2.0.co;2 (2005).
Grinstein, S. et al. Demonstration of Epstein-Barr virus in carcinomas of various sites. Cancer research 62, 4876–4878 (2002).
Mazouni, C. et al. Epstein-Barr virus as a marker of biological aggressiveness in breast cancer. British journal of cancer 104, 332–337, 10.1038/sj.bjc.6606048 (2011).
Joshi, D. & Buehring, G. C. Are viruses associated with human breast cancer? Scrutinizing the molecular evidence. Breast cancer research and treatment 135, 1–15, 10.1007/s10549-011-1921-4 (2012).
Rickinson, A. B. & Kieff, E. in Virology Vol. 2, 2397–2446. (Lippincott-Raven Publishers, 1996).
Lawson, J. S. et al. Presence of mouse mammary tumour-like virus gene sequences may be associated with morphology of specific human breast cancer. Journal of clinical pathology 59, 1287–1292, 10.1136/jcp.2005.035907 (2006).
Ford, C. E., Faedo, M., Crouch, R., Lawson, J. S. & Rawlinson, W. D. Progression from normal breast pathology to breast cancer is associated with increasing prevalence of mouse mammary tumor virus-like sequences in men and women. Cancer research 64, 4755–4759, 10.1158/0008-5472.CAN-03-3804 (2004).
Melana, S. M., Holland, J. F. & Pogo, B. G. Search for mouse mammary tumor virus-like env sequences in cancer and normal breast from the same individuals. Clinical cancer research: an official journal of the American Association for Cancer Research 7, 283–284 (2001).
Fukuoka, H., Moriuchi, M., Yano, H., Nagayasu, T. & Moriuchi, H. No association of mouse mammary tumor virus-related retrovirus with Japanese cases of breast cancer. Journal of medical virology 80, 1447–1451, 10.1002/jmv.21247 (2008).
Motamedifar, M., Saki, M. & Ghaderi, A. Lack of association of mouse mammary tumor virus-like sequences in Iranian breast cancer patients. Medical principles and practice: international journal of the Kuwait University, Health Science Centre 21, 244–248, 10.1159/000334572 (2012).
Adams, A. Replication of latent Epstein-Barr virus genomes in Raji cells. Journal of virology 61, 1743–1746 (1987).
Liu, P. et al. Direct sequencing and characterization of a clinical isolate of Epstein-Barr virus from nasopharyngeal carcinoma tissue by using next-generation sequencing technology. Journal of virology 85, 11291–11299, 10.1128/JVI.00823-11 (2011).
Moore, P. S. & Chang, Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nature reviews. Cancer 10, 878–889, 10.1038/nrc2961 (2010).
Callahan, R. & Smith, G. H. MMTV-induced mammary tumorigenesis: gene discovery, progression to malignancy and cellular pathways. Oncogene 19, 992–1001, 10.1038/sj.onc.1203276 (2000).
Katz, E. et al. MMTV Env encodes an ITAM responsible for transformation of mammary epithelial cells in three-dimensional culture. The Journal of experimental medicine 201, 431–439, 10.1084/jem.20041471 (2005).
Ross, S. R. et al. An immunoreceptor tyrosine activation motif in the mouse mammary tumor virus envelope protein plays a role in virus-induced mammary tumors. Journal of virology 80, 9000–9008, 10.1128/JVI.00788-06 (2006).
Shair, K. H. et al. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) and LMP2A function cooperatively to promote carcinoma development in a mouse carcinogenesis model. Journal of virology 86, 5352–5365, 10.1128/JVI.07035-11 (2012).
Curran, J. A., Laverty, F. S., Campbell, D., Macdiarmid, J. & Wilson, J. B. Epstein-Barr virus encoded latent membrane protein-1 induces epithelial cell proliferation and sensitizes transgenic mice to chemical carcinogenesis. Cancer research 61, 6730–6738 (2001).
Bieging, K. T., Swanson-Mungerson, M., Amick, A. C. & Longnecker, R. Epstein-Barr virus in Burkitt's lymphoma: a role for latent membrane protein 2A. Cell cycle 9, 901–908 (2010).
Franco, A. T. et al. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer research 68, 379–387, 10.1158/0008-5472.CAN-07-0824 (2008).
Peek, R. M., Jr & Crabtree, J. E. Helicobacter infection and gastric neoplasia. The Journal of pathology 208, 233–248, 10.1002/path.1868 (2006).
JJ, B. Possible Relationship of the Estrogenic Hormones, Genetic Susceptibility and Milk Influence in the Production of Mammary Cancer in Mice. Cancer research 2, 710–721 (1942).
Rickinson, A. B. & Kieff, E. in Fields Virology Vol. 2, (eds Knipe D. M., & Howley P. M.) Ch. 68B, 2655–2700. (Lippincott-Raven Publishers, 2007).
Subramanian, C., Cotter, M. A., 2nd & Robertson, E. S. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nature medicine 7, 350–355, 10.1038/85499 (2001).
Chu, P. G., Chang, K. L., Chen, Y. Y., Chen, W. G. & Weiss, L. M. No significant association of Epstein-Barr virus infection with invasive breast carcinoma. The American journal of pathology 159, 571–578, 10.1016/s0002-9440(10)61728-2 (2001).
Fina, F. et al. Frequency and genome load of Epstein-Barr virus in 509 breast cancers from different geographical areas. British journal of cancer 84, 783–790, 10.1054/bjoc.2000.1672 (2001).
Mok, M. T., Lawson, J. S., Iacopetta, B. J. & Whitaker, N. J. Mouse mammary tumor virus-like env sequences in human breast cancer. International journal of cancer. Journal international du cancer 122, 2864–2870, 10.1002/ijc.23372 (2008).
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J. & Wheeler, D. L. GenBank. Nucleic acids research 31, 23–27 (2003).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. Journal of molecular biology 215, 403–410, 10.1016/S0022-2836(05)80360-2 (1990).
Epi Info™, a database and statistics program for public health professionals. (CDC, Atlanta, GA, 2011).
Acknowledgements
This work was funded by National Council of Science and Technology: Fondo SEP CONACyT 80400 to EM Fuentes-Pananá, sectorial CONACyT 69450 to J Torres; and Fondo de Investigación en Salud (FIS/IMSS/PROT/C2007/023) to EM Fuentes-Pananá. This paper constitutes a partial fulfillment of the Graduate Program of Doctor Degree in Biomedical Sciences, National Autonomous University of Mexico, Mexico City, Mexico, to A Morales-Sánchez. It also constitutes a partial fulfillment of the Graduate Program in Biological Sciences, National Autonomous University of Mexico, Mexico City, Mexico to JLE Martínez-López. A Morales-Sánchez and JLE Martínez-López also acknowledge the scholarship and financial support provided by the National Council of Science and Technology (CONACyT), National Autonomous University of Mexico and IMSS. We want to thank Dr. Mario Vega, (Oncology Hospital at the National Medical Center “XXI Century”, IMSS, Mexico City Mexico) and Dr. Paul D. Ling (Baylor College of Medicine, Houston TX, USA) for the reagents donated to accomplish this study.
Author information
Authors and Affiliations
Contributions
A.M.S. is first author on this manuscript. E.M.F.P. is senior author on this manuscript. E.M.F.P. conceived and designed experiments. A.M.S., T.M.M., J.L.E.M.L. and P.H.S. performed the experiments. A.M. carried out all samples histopathological examination. Y.A.L., J.T. and E.M.F.P. coordinated the study. A.M.S., J.E.M.L. and E.M.F.P. analyzed the data, wrote the main manuscript text and prepare figures 1–5. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Morales-Sánchez, A., Molina-Muñoz, T., Martínez-López, J. et al. No association between Epstein-Barr Virus and Mouse Mammary Tumor Virus with Breast Cancer in Mexican Women. Sci Rep 3, 2970 (2013). https://doi.org/10.1038/srep02970
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02970
This article is cited by
-
The association of infectious mononucleosis and invasive breast cancer in The Health of Women (HOW) Study®
Breast Cancer (2022)
-
Prevalence of Human Papillomavirus and Mouse Mammary Tumor Virus Like DNAs in Tumors from Moroccan Breast Cancer Patients
Indian Journal of Gynecologic Oncology (2022)
-
Does mouse mammary tumor-like virus cause human breast cancer? Applying Bradford Hill criteria postulates
Bulletin of the National Research Centre (2020)
-
Co-presence of human papillomaviruses and Epstein–Barr virus is linked with advanced tumor stage: a tissue microarray study in head and neck cancer patients
Cancer Cell International (2020)
-
Prevalence of Mouse Mammary Tumor Virus (MMTV)-like sequences in human breast cancer tissues and adjacent normal breast tissues in Saudi Arabia
BMC Cancer (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.