Abstract
The introduction of Candida albicans into cefoperazone-treated mice results in changes in bacterial community reassembly. Our objective was to use high-throughput sequencing to characterize at much greater depth the specific changes in the bacterial microbiome. The colonization of C. albicans significantly altered bacterial community reassembly that was evident at multiple taxonomic levels of resolution. There were marked changes in the levels of Bacteriodetes and Lactobacillaceae. Lachnospiraceae and Ruminococcaceae, the two most abundant bacterial families, did not change in relative proportions after antibiotics, but there were marked genera-level shifts within these two bacterial families. The microbiome shifts occurred in the absence of overt intestinal inflammation. Overall, these experiments demonstrate that the introduction of a single new microbe in numerically inferior numbers into the bacterial microbiome during a broad community disturbance has the potential to significantly alter the subsequent reassembly of the bacterial community as it recovers from that disturbance.
Similar content being viewed by others
Introduction
Exposure to broad spectrum antibiotics, such as cephalosporins, causes marked shifts in bacterial community structure, including decreased numbers, reduced diversity, changes in membership and changes in evenness1,2,3,4,5. After cessation of antibiotics, bacterial communities in the GI tract will generally reassemble to a pre-antibiotic structure. However, complete recovery is not always the case1,2,3,4,5. The factors that drive community reassembly have not been completely elucidated. Colonization of the microbiome by a new microbe can occur during the period of diminished colonization resistance that immediately follows antibiotic therapy. This "invasion" of the community has the potential to alter reassembly dynamics through stimulation of mucosal inflammation, alteration of microbial metabolic networks, interference with quorum sensing and alteration of adhesion6,7,8.
Yeast are naturally resistant to antibacterial antibiotics and the contribution of yeast to modulating post-antibiotic bacterial community reassembly is not well understood. Interplay between yeast and bacteria in polymicrobial communities in the host and environment have been reported9,10,11,12, consistent with the concept that growth of yeast populations in the microbiota has the potential to alter bacterial community reassembly. On a healthy mucosa, yeast (e.g., Candida albicans) are kept at low levels or excluded by yet-to-be-defined mechanisms of colonization resistance mediated by the indigenous bacterial microbiota13,14,15,16,17,18,19. Antibiotics, notably broad spectrum antibiotics, can disrupt colonization resistance, leading to yeast overgrowth (either as an indigenous "bloom" or by exogenous invasion into the disrupted microbiota)20,21. The cessation of antibiotics can quite often reverse the pathogenic process attributed to high yeast numbers on the mucosa. However, there is an ever-increasing body of research documenting that changes in the bacterial microbiota alone can alter mucosal biology22,23,24,25,26,27,28. Thus, there remains the question of whether the bloom or invasion of yeast in a disrupted microbiome can alter bacterial community reassembly following broad disturbance and whether this alteration will modify host mucosal biology.
We have previously addressed this second question about modification of host mucosal biology by yeast. We demonstrated that the presence of C. albicans in the GI microbiota during recovery from treatment with a third generation cephalosporin can alter host mucosal immunity at a distal site, rendering mice susceptible to developing allergic airway disease24,25. Since yeast in the absence of antibiotic disturbance did not render the host susceptible, nor did antibiotics alone, these data raised the possibility that one mechanism may involve alteration of bacterial community reassembly by the introduced yeast. We subsequently demonstrated, using selective culture and 16S rRNA gene clone libraries, that there were changes in bacterial community reassembly when C. albicans was present29. Our primary objective in this current study was to characterize at much greater depth the specific changes in the bacterial community via 454 pyrosequencing of 16S rRNA gene amplicon libraries, similar to our previous studies analyzing intestinal conventionalization dynamics in mice30. Since intestinal inflammation can alter the bacterial microbiota, we also sought to determine whether intestinal inflammation was playing a significant role in this process.
Results
Disturbing the indigenous bacterial community by a broad spectrum antibiotic breaks colonization resistance and allows long-term colonization of the ileum and cecum by C. albicans CHN1
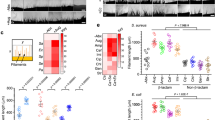
C57BL/6 mice were treated with cefoperazone in their drinking water for one week (days −7 to 0), then switched to sterile drinking water and inoculated with C. albicans CHN1 by oral gavage at day 0. The groups of mice were subsequently analyzed for bacterial microbiome changes and C. albicans colonization at days 7 and 21 post-antibiotic treatment (Figs. 1A & 1B). Cefoperazone reduced total bacterial numbers in the intestine by 10,000 fold during the week the mice were drinking the antibiotic water (determined by both culture and total 16S rRNA gene qPCR, data not shown). Upon cessation of the antibiotic, total bacterial numbers returned to pre-antibiotic levels by 72 hours (approx. 1010 culturable bacteria per g cecal tissue). Neither untreated mice nor those whose microbiome had been disturbed by cefoperazone had detectable C. albicans in their ileum or cecum at days 7 and 21 (Figs. 1A & 1B). When mice were gavaged with C. albicans in the absence of microbiome disturbance, less than half of the mice had detectable C. albicans levels at day 7 and almost none at day 21. In sharp contrast, all of the gavaged mice with a disturbed microbiome had detectable C. albicans colonization at day 7 and the colonization levels did not significantly change through day 21. We have followed mice out through 90 days and C. albicans levels in C. albicans-gavaged mice remain similar to those seen at day 21 (data not shown). Thus, the indigenous bacterial microbiota exerts strong competitive exclusion against invasion by the yeast C. albicans; however, disturbance of the community by cefoperazone breaks colonization resistance and allows C. albicans to colonize the intestinal microbiome at levels far inferior to those of the bacteria (>1010 bacteria vs. 104 yeast).
The effect of C. albicans colonization on bacterial community structure.
C57BL/6 mice were treated with cefoperazone in their drinking water for one week (days -7 to 0), switched to sterile drinking water and then inoculated with C. albicans CHN1 by oral gavage at day 0. The groups of mice were then analyzed for bacterial microbiome changes and C. albicans colonization at days 7 and 21 (n = 8–12 mice per treatment per time point). Panels A and B demonstrate the growth of C. albicans from the ileum and cecum at day 7 and 21 (L.O.D. of 101.7). The inverse-Simpson diversity index was utilized to quantify the diversity of the cecal communities at day 7 and day 21 (panel C). Significance was tested using 1-way ANOVA (p < 0.05). Rarefaction analysis was also employed to view what idealized collector's curve based on the data would look like at day 7 and day 21 (panel D). The mean curve of 8–10 rarefaction curves is displayed as a bold line and the dotted lines represent the standard error. Panel E is a canonical correspondence analysis (CCA) plot of both day 7 and 21 normalized microbial composition data where the data has been constrained by treatment and time. To better visualize the group clusters, each data point is connected to its group centroid.
After disturbance of the GI microbiota, colonization by C. albicans CHN1 at low numbers can alter the reassembly of the bacterial community
To assess the effects of C. albicans CHN1 colonization on the reassembly of the intestinal bacterial community after broad disturbance, we used 454 pyrosequencing of V3–V5 16S rRNA gene amplicon libraries generated from total cecal mucosal DNA at days 7 and 21 post-disturbance. Sequence data were binned into operational taxonomic units (OTU), based on 97% similarity, as described in the methods.
Two metrics of change in bacterial diversity were calculated for each of the treatment groups at day 7 and day 21: inverse-Simpson values and rarefaction curves (Figs. 1C & 1D). The inoculation of C. albicans into an undisturbed community did not significantly affect diversity. However, there was a significant decrease in the diversity of the bacterial community of the cecum at day 7 post-disturbance compared to untreated mice. The inoculation of C. albicans into a disturbed community further decreased community diversity at day 7 post-antibiotic, although the additional loss of diversity did not score as statistically significant. By day 21, the diversity of both the groups that had suffered a microbiota disturbance had largely recovered to that of the untreated groups, although the diversity of the group that was disturbed and gavaged with C. albicans still trended lower.
Next, the bacterial community structure of the experimental groups was assessed using canonical correspondence analysis (CCA). This is a method of direct gradient analysis commonly used in ecology37,38 where the community data matrix is subjected to a weighted linear regression on the constraining variables and a correspondence analysis performed on the fitted values. This both focuses the data on the question of interest and creates a testable model for assessment of significant effects. In our study, the variation was constrained to treatment group and time (Fig. 1E). The communities in the untreated (undisturbed) mice shifted very little over the course of the experiments (day 7 vs. day 21). The addition of C. albicans to the undisturbed state had very little effect on the bacterial communities at day 7 or day 21. In contrast, disturbance of the bacterial community by cefoperazone treatment resulted in a significant shift (p < 0.005) away from the untreated state, even seven days after the cessation of antibiotics. The introduction of C. albicans into this disturbed bacterial community resulted in a community structure that was not only significantly different from that of untreated mice, but was also significantly different from mice treated with antibiotics alone. Between days 7 and 21 post-antibiotic, the bacterial intestinal communities in both of the disturbed groups changed to become more similar to that in the undisturbed group (i.e., reassembly). However, the bacterial microbiota of mice treated with antibiotics (disturbed) and of antibiotic-disturbed with C. albicans gavage remained significantly different from each other.
Colonization of the ileum and cecum by C. albicans elicits a host response but does not induce intestinal inflammation
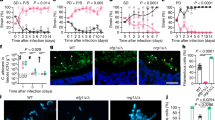
Mice were examined at days 7 and 21 post-antibiotic for intestinal inflammation (histology) and at day 21 for changes in intestinal and mesenteric lymph node gene expression by high-throughput qPCR. When C. albicans was introduced into an undisturbed microbiota (Fig. 2B), very few genes showed any significant change in expression, while disturbance of the microbiota caused a number of genes to be significantly altered in expression (Fig. 2C). The addition of C. albicans to a disturbed microbiota (Fig. 2D) resulted in changes in gene expression not seen in the other groups. The effects of these treatments on the expression of immune modulatory molecules in the mesenteric lymph nodes followed a similar trend (Fig. 2A). However, despite the active differential regulation of genes, there was no evidence of inflammation in histological sections of the small intestine (defined by absence of leukocyte infiltrates, edema or changes in epithelium structure) and no histological evidence of hyphal invasion (Fig. 3). We have reported previously that C. albicans colonization does not induce cecal inflammation25,29.
The response of the host to microbiota disturbance and C. albicans colonization.
(A) Panel A depicts a heatmap of gene expression in the ileum and mesenteric lymph nodes as determined by multiplex qPCR array. For gene identities see Table S1. Fold changes are depicted relative to the undisturbed state. Panels B–D depict genes in which a statistically significant difference occurred are displayed for each of the treatment groups (B) C. albicans alone, (C) disturbed microbiota, or (D) disturbed microbiota colonized with C. albicans. Significant differences in gene expression were determined using SAM (p < 0.05, false discovery rate set at zero (q = 0.00)) are displayed for each of the treatment groups) as described in Methods.
Photomicrograph of the ileum of mice in the treatment groups (Day 21).
Figure 3 displays Hematoxylin & Eosin stained histology of the small intestine from mice that were (A) Undisturbed (untreated), (B) C. albicans only, (C) disturbed microbiota (Cefoperazone only), or (D) disturbed microbiota colonized with C. albicans (disturbed plus C. albicans). Mice were treated as described in Fig. 1 and the manuscript text.
Introduction of C. albicans into a disturbed bacterial community can alter the diversity and membership of the community
We next characterized the bacterial lineages that shifted during disturbance and recovery, by using RDP-based taxonomic assignment of the bacterial OTUs (97% identity), as described in the methods. At day 7, significant phylum-level shifts were evident in both the antibiotic-disturbed community and the community of antibiotic-disturbed with C. albicans gavage (Fig. 4A). There were significant decreases in both the Bacteroidetes and the Synergistetes. The dominant phylum was the Firmicutes, which remained unchanged no matter the treatment.
Phylum and family taxonomic membership of day 7 treatment groups.
OTUs were classified according to the RDP taxonomy and rank abundance graphs constructed for the phylum and family-level taxonomies. Panel A is the phylum-level classification for the day 7 undisturbed (red), undisturbed plus C. albicans (orange), disturbed (green) or disturbed plus C. albicans (blue) groups. Panels B–E are the family-level taxonomic distributions. X-axis ordering of the rank abundance graphs is based on the relative distribution in the undisturbed community. Data are mean ± SEM; n = 8–10 mice per group from 3 separate experiments.
Analysis at a deeper taxonomic level, revealed that there were a number of marked changes in bacterial community membership at the family-level (Figs. 4B–4E). Both the Rikenellaceae and the Porphyromonadaceae, among the top 5 most abundant families in the undisturbed treatment group, were all but non-existent in either of the disturbed treatment groups. In contrast, bacteria from less abundant families (in untreated mice) bloomed in the mice with the disturbed microbiota.
Furthermore, there were notable changes at day 7 within the Lachnospiraceae (Fig. 6A) and Ruminococcaceae (Fig. 6B). These two bacterial families were (1) the most abundant in all the experimental groups and (2) did not appear to change in relative proportions after antibiotics. At the genus-level, the disturbance of the microbiota resulted in a marked shift in the dominant genera within these two familes (Fig. 6). In undisturbed communities, Lachnobacterium, Marvinbryantia and Syntrophococcus constituted approximately 80% of the Lacnospiraceae. However, after antibiotic treatment, they accounted for <50% of the population at day 7. There were marked blooms of the rare Lachnospiraceae genera Coprococcus, Anaerosporobacter, Dorea and Robinsoniella in antibiotic-only mice. In contrast, C. albicans colonization of the disturbed community was associated with the rapid recovery of Lachnobacterium and Syntrophococcus to undisturbed levels, outgrowth of Roseburia and Robinsoniella and reduction in Coprococcus and Dorea levels (Fig. 6). In the Ruminococcaceae, disturbance had very little effect on the dominant genera within the community; however, the presence of C. albicans in the disturbed community resulted in almost a complete elimination of Oscillibacter from the community.
Phylum and family taxonomic membership of day 21 treatment groups.
OTUs were classified according to the RDP taxonomy and rank abundance graphs constructed for the phylum and family-level taxonomies. Panel A is the phylum-level classification for the day 21 undisturbed (red), undisturbed plus C. albicans (orange), disturbed (green) or disturbed plus C. albicans (blue) groups. Panels B–E are the family-level taxonomic distributions. X-axis ordering of the rank abundance graphs is based on the relative distribution in the undisturbed community. The arrows in panels D and E highlight the Lactobacillaceae, which remain significantly reduced at day 21 following cefoperazone treatment. Data are mean ± SEM; n = 8–10 mice per group from 3 separate experiments.
Genus-level taxonomy of the five most abundant families (Day 7).
The top 5 most abundant families (Lachnospiraceae, Ruminococcaceae, Rikenellaceae, Porphyromonadaceae and Incerte Sedis XIV, respectively) were further classified at a genus-level taxonomy (panels A–E). Data are mean; n = 8–10 mice per group pooled from 3 separate experiments.
A similar analysis was next carried out at the phylum, family and genus-level at day 21 post-disturbance. The bacterial community structure of the antibiotic-treated mice still had not recovered to a pre-antibiotic state (Fig. 5). Most notably, the levels of the Bacteroidetes phylum was 100 fold lower level than in undisturbed levels (Fig. 5A). However, the presence of low levels of C. albicans within the disturbed community was associated with higher Bacteroidetes levels than in antibiotic only treated mice, similar to untreated mice (Fig. 5A). The blooms in low abundance bacteria that were seen at day 7 were no longer evident at day 21. Relative levels of six of the eleven most dominant families were still markedly depressed in antibiotic-only mice at day 21. In contrast, the presence of C. albicans during the recovery process was associated with ten of the eleven most dominant bacterial families being found at relative levels similar to that found in untreated (undisturbed) mice. The one notable exception was the very low levels of the Lactobacillaceae (denoted by arrows, Figs. 5D & 5E) in both the groups that had undergone an antibiotic-disturbance. In contrast to day 7, there were no longer genus-level differences in the Lachnospiraceae and Ruminococcaceae at day 21 (Fig. 7), largely indicating that the Lachnospiraceae populations had finally recovered fully from cefoperazone treatment.
Genus-level taxonomy of the five most abundant families (Day 7).
The top 5 most abundant families (Lachnospiraceae, Ruminococcaceae, Rikenellaceae, Porphyromonadaceae and Incerte Sedis XIV, respectively) were further classified at a genus-level taxonomy (panels A–E). Data are mean; n = 8–10 mice per group pooled from 3 separate experiments.
Discussion
These experiments demonstrate that the introduction of a single new microbe (the yeast C. albicans CHN1) in numerically inferior numbers into the bacterial microbiome during a broad community disturbance can significantly alter the subsequent reassembly of the bacterial community as it recovers from that disturbance. This occurs in the absence of overt intestinal inflammation (leukocyte infiltrates, edema or changes in epithelium structure) but is associated with changes in host gene expression in the intestinal epithelium. We have previously demonstrated that the recovery of the microbiota in the presence of C. albicans leads to a state of systemic hyperresponsiveness24,25. The effects of the antibiotics and yeast were localized to the GI tract because (1) cefoperazone is poorly absorbed when delivered in the drinking water, (2) we have been able to demonstrate that oral treatment with cefoperazone is unable to clear a cefoperazone-sensitive bacteria from the lungs of mice (data not shown) and (3) yeast are not introduced into the lungs following oral gavage24,25. We propose that our current studies provide additional insight into potential pathways by which colonization with a numerically inferior organism after antibiotics can have a disproportionate effect on the immune system without inducing intestinal inflammation: by affecting the reassembly of the bacterial microbiome.
Our study demonstrates that C. albicans can colonize the mouse cecum and ileum without causing overt intestinal inflammation but the presence of C. albicans is associated with changes in bacterial community reassembly. Although C. albicans, the introduced microbe, is not a normal member of the mouse microbiota, it is known to first colonize humans within a few weeks after birth39. In mice, C. albicans is excluded by the normal microbiota; however, colonization can take place if the GI microbiota is disturbed. Interestingly, while C. albicans causes a mild gastritis in the stomach, the fungi colonizes other parts of the lower GI tract with no evidence of local inflammation31. A diverse and abundant fungal microbiome, as detected by molecular techniques has been reported previously, using mice from both a specific pathogen-free (SPF) source and from a colony with a restricted bacterial flora (RF mice)40. In a recently published study with Dectin-1 knockout and wildtype mice, high levels of indigenous fungi were reported in wildtype laboratory mice, especially C. tropicalis and accounted for 65% of the fungi detected by ITS1-2 amplicon pyrosequencing, although no culture data was presented for this yeast that can be readily cultured on Sabouraud Dextrose Agar and identified on Chromagar41. In our laboratory, as we have reported here, we have observed that SPF mice routinely lack culturable fungi. Treatment with a broad spectrum antibiotic, such as cefoperazone, can result in the culture of fungi from intestinal samples on SDA, such as C. tropicalis25,29,31, but after cessation of cefoperazone, these yeast are rarely detected in the mice. The systemic hyperresponsiveness of Candida-colonized mice to challenge with an aeroallergen24,25 indicates that intestinal C. albicans colonization can alter the immune response of the host. The recent report demonstrating a role for Candida species in driving colitis in Dectin-1 knockout and wildtype mice41 further underscores the potential of the fungal microbiome to alter host immunity and inflammation.
The current work also highlights that members of the Porphyromonadaceae, Rikenellaceae, Lachnospiraceae and Lactobacillaceae bacterial families are some of the most susceptible to perturbations in reassembly following cefoperazone treatment (+/− C. albicans). While it is unknown how the mouse microbiota prevents C. albicans colonization, previous studies have demonstrated the ability of C. albicans to compete with bacteria (particularly amongst members of the Firmicutes)42. Interactions between C. albicans and other phyla, such as the Bacteroidetes, are not well investigated, although it has been suggested that Lactobacillus species are critical15,43. To induce a disturbance in the microbial community and enable C. albicans colonization, cefoperazone was used. This is a poorly absorbed third-generation cephalosporin with excellent activity against anaerobic bacteria44. It can facilitate gastric colonization by C. albicans31. Cefoperazone has also been shown to be able to disturb the mouse cecal microbiota for greater than six weeks after washout if the mice are housed in isolation1. This delay in reassembly is in contrast to the observation that a cecal microbiota community can completely reassemble within seven days following transfer from a conventional to a germ-free mouse, despite a complete destruction of community structure during the first few days after transfer30. In addition, cefoperazone-treated mice will completely and quickly reassemble their cecal microbiota if allowed to recover in the presence of an untreated mouse1. While currently of unknown significance, the association between C. albicans colonization and altered Lachnospiraceae composition is intriguing in light of the report that Lachnospiraceae communities can be important in colonization resistance against enteric bacterial pathogens45.
We have observed that the intestinal microbiota shift caused by the introduction of C. albicans into cefoperazone-treated mice is not associated with mucosal inflammation, but it is also not a neutral event for the host. The pattern of genes down-regulated in mice with a disturbed microbiota and C. albicans colonization is consistent with an environment where the host is less responsive to microbial signals. Down-regulated genes of interest included Ffar2 (Gpr43)46,47,48, Nod149,50, Gpr3551,52 and the TLRs26,53. These are all well-characterized as environmental sensors that play important roles in inflammation. At the same time, there is a decrease in the expression of various anti-microbial peptides and tight-junction proteins that might render the host less capable of maintaining microbial homeostasis54,55,56. Thus, it is possible that the communication between the host and the GI microbiota has been disrupted but not to the breaking point to create intestinal inflammation. It also remains possible that, despite a lack of intestinal inflammation, the mild inflammation in the stomach may be influencing gene expression in the cecum, although it is unclear how this signaling would take place. Another possibility is that formation of discrete foci of yeast invasion and inflammation is occurring in the intestine but is undetectable at the level of resolution used in this study. Finally, active suppression of host responsiveness by C. albicans or other members of the microbiota remains a possibility because it has been previously demonstrated that C. albicans can produce prostaglandins and related oxylipins57,58,59. In summary, we have provided a proof-of-concept study that low-level colonization and/or blooms of yeast within the microbiome following antibiotic therapy can potentially have unintended consequences on bacterial microbiota reassembly and host mucosal immunity.
Methods
Animals and housing
Female C57BL/6 mice (Jackson Laboratories, Indianapolis, IN) were housed 5 mice to a cage under specific-pathogen-free conditions in enclosed filter-top cages. The experiments (at days 7 and 21 post-treatment) were performed on 3 separate occasions. Food and sterile water were given ad libitum. Food remained constant throughout the experiment to minimize the effect of diet on the microbiota. Mice were maintained on grates by the Unit for Laboratory Animal Medicine (ULAM) at the University of Michigan (Ann Arbor, MI) to prevent coprophagy. All protocols were approved by the University of Michigan University Committee on Use and Care of Animals (UCUCA).
Antibiotic treatment
Cefoperazone (0.5 mg/ml) (Sigma-Aldrich, St. Louis, MO) was administered orally to mice ad libitum in drinking water. Antibiotic treatment was continued for 7 days (days −7 to day 0) prior to C. albicans colonization. After 7 days, antibiotic containing drinking water was replaced with sterile water.
C. albicans GI inoculation
C. albicans strain CHN1 (a human clinical isolate) was grown as previously described31, diluted to 2 × 108 CFU/ml in sterile nonpyrogenic saline and 107 CFU in 50 μl gavaged using a 24-gauge feeding needle attached to a 1-ml syringe mounted on a Stepper repetitive pipette (Tridak, Brookfield, CT) to deliver an equivalent amount of inoculum to each mouse. The inoculums were plated on Sabouraud dextrose agar (SDA) to verify the number of colony forming units (CFU) delivered.
Tissue isolation
Following euthanasia, the GI tract was sterilely removed and the cecum separated from the colon and small intestine. A 1 cm2 tissue strip from the cecal tip was removed, rinsed in sterile 1× PBS and divided for DNA and RNA isolation. The remaining portion of the cecum was cut open along the greater curvature and rinsed in sterile 1× PBS. Thin slices along the outer portion were removed and fixed overnight in 10% Neutral Buffered Formalin for histological analysis. The remaining tissue was homogenized in sterile water using a Tissuemiser (Fisher Scientific). Aliquots of the homogenates were plated in 10-fold serial dilutions on Sabouraud dextrose agar (Difco). For the small intestine, the distal half of the organ was separated, cut open longitudinally and rinsed in sterile 1× PBS. A 1 cm2 tissue strip was taken from the middle and divided for DNA and RNA isolation. The distal portion of the remaining tissue was prepared for histological analysis using the "Swiss-roll" technique whereby the tissue is rolled and fixed overnight in 10% Neutral Buffered Formalin, which more readily allows visualization of the entire length of the intestinal sample. The proximal portion of the remaining tissue was homogenized in sterile water using a Tissuemiser (Fisher Scientific). Aliquots of the homogenates were plated in 10-fold serial dilutions on Sabouraud dextrose agar (Difco) to determine levels of C. albicans colonization.
DNA extraction
Genomic DNA was extracted from tissue homogenized in 360 μl ATL buffer (Qiagen DNeasy Blood & Tissue kit) using a modified protocol previously demonstrated to isolate bacterial DNA31.
454-prosequencing
The V3–V5 region of the bacterial 16S rRNA gene was targeted using barcoded primer sets corresponding to 357F and 929R. These primers sets were originally developed by the Broad Institute. The PCR conditions were 95°C for 2 min., followed by 30 cycles of (95°C 20 sec, 50°C 30 sec, 72°C 5 min), holding at 4°C. Quality control and sequencing was carried out at the University of Michigan, using the Roche 454 GS-FLX Titanium platform.
16S rRNA gene data analysis
Sequence data were processed and analyzed using the software mothur v. 1.2132 according to the protocol provided (http://www.mothur.org). The operational taxonomic units (OTUs) were binned at 97% similarity (to create shared community file and the phylotyped, genus-level grouping, file). For the purposes of this study, we focused on OTUs that were present at greater than 0.5% of the sample population. These files, along with the file containing the taxonomic information for the OTUs, were imported and further analyzed in R. The R-package vegan33 was used for diversity analyses and ordinations, as well as a custom R script for sorting classification results into tables (ClassifierSorter v.2). Classification of OTUs was carried out using the implementation of the RDP Classifier and RDP taxonomy training set 734,35 contained in mothur. The number of sequences in each sample group can be found in Supplemental Table S2.
Quantification of mRNA levels using high-throughput quantitative PCR
RNA was isolated from each tissue using TRIzol (Ambion) and the RNeasy Mini Kit (Qiagen). RNA concentration was determined using a Nanodrop instrument (Thermo Scientific). Complementary DNA (cDNA) was generated using the RT2 First Strand Kit (Qiagen). Expression levels of genes under study were determined using Mouse RT2Profiler PCR assays, custom-made to contain replicate sets of 48 primer pairs (Qiagen). Each well of the replicate sets was loaded with 2.5 ng of cDNA reaction product. Each card was run on a LightCycler 480 Real-Time PCR system (Roche). The relative RNA expression levels were inferred from the Ct values. The geometric mean of ß-actin and GAPDH levels was used as the internal control for the generation of ΔCt values. The heatmap was generated in R using the package gplots36.
Statistics
A standard one-way ANOVA with a Tukey post-test was used to determine whether statistical differences existed in C. albicans CFU counts and in diversity metrics (p < 0.05 considered significant). The R implementation of the Statistical Analysis of Microarrays (SAM) (R-package samr) was used to determine statistical differences in gene expression with significance (p < 0.05) being set for q = 0.00 (q-value of a gene equals the minimum estimated false discovery rate at which it appears significant). Differences in microbial community centroids were determined using a permutational MANOVA (function adonis() in the R-package vegan). The significance of constraints in constrained ordinations was tested using an ANOVA-like permutation test (function anova.cca in R-package vegan). All statistical analyses were performed in R or Prism v5.
References
Antonopoulos, D. A. et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun 77, 2367–2375 (2009).
Dethlefsen, L., Huse, S., Sogin, M. L. & Relman, D. A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6, e280 (2008).
Dethlefsen, L. & Relman, D. A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108 Suppl 1, 4554–4561 (2011).
Jernberg, C., Lofmark, S., Edlund, C. & Jansson, J. K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. The ISME journal 1, 56–66 (2007).
Lofmark, S., Jernberg, C., Jansson, J. K. & Edlund, C. Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. The Journal of antimicrobial chemotherapy 58, 1160–1167 (2006).
Antunes, L. C., Davies, J. E. & Finlay, B. B. Chemical signaling in the gastrointestinal tract. F1000 biology reports 3, 4, 10.3410/B3-4 (2011).
Round, J. L. & Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nature reviews. Immunology 9, 313–323 (2009).
Willing, B. P., Russell, S. L. & Finlay, B. B. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nature reviews. Microbiology 9, 233–243 (2011).
McAlester, G., O'Gara, F. & Morrissey, J. P. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. Journal of medical microbiology 57, 563–569 (2008).
Minervini, F. et al. Lactic acid bacterium and yeast microbiotas of 19 sourdoughs used for traditional/typical italian breads: interactions between ingredients and microbial species diversity. Applied and environmental microbiology 78, 1251–1264 (2012).
Shirtliff, M. E., Peters, B. M. & Jabra-Rizk, M. A. Cross-kingdom interactions: Candida albicans and bacteria. FEMS microbiology letters 299, 1–8 (2009).
Zijnge, V. et al. Oral biofilm architecture on natural teeth. PloS one 5, e9321 (2010).
Hummel, R. P., Oestreicher, E. J., Maley, M. P. & Macmillan, B. G. Inhibition of Candida albicans by Escherichia coli in vitro and in the germfree mouse. J Surg Res 15, 53–58 (1973).
Kennedy, M. J. Inhibition of Candida albicans by the anaerobic oral flora of mice in vitro. Sabouraudia 19, 205–208 (1981).
Kennedy, M. J. & Volz, P. A. Ecology of Candida albicans gut colonization: inhibition of Candida adhesion, colonization and dissemination from the gastrointestinal tract by bacterial antagonism. Infect Immun 49, 654–663 (1985).
Naglik, J. R., Fidel, P. L., Jr & Odds, F. C. Animal models of mucosal Candida infection. FEMS Microbiol Lett 283, 129–139 (2008).
van der Waaij, D. Colonization resistance of the digestive tract--mechanism and clinical consequences. Nahrung 31, 507–517 (1987).
van der Waaij, D. & Berghuis, J. M. Determination of the colonization resistance of the digestive tract of individual mice. J Hyg (Lond) 72, 379–387 (1974).
Van der Waaij, D. & Van der Waaij, B. D. The colonization resistance of the digestive tract in different animal species and in man; a comparative study. Epidemiol Infect 105, 237–243 (1990).
Calderone, R. A. & Clancy, C. J. Candida and candidiasis. 2nd edn, (ASM Press, 2012).
Mandell, G. L., Bennett, J. E. & Dolin, R. Mandell, Douglas and Bennett's principles and practice of infectious diseases. 7th edn, (Churchill Livingstone/Elsevier, 2010).
Hill, D. A. et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med 18, 538–546 (2012).
Ivanov, II et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Noverr, M. C., Falkowski, N. R., McDonald, R. A., McKenzie, A. N. & Huffnagle, G. B. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen and interleukin-13. Infect Immun 73, 30–38 (2005).
Noverr, M. C., Noggle, R. M., Toews, G. B. & Huffnagle, G. B. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect Immun 72, 4996–5003 (2004).
Round, J. L. et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977 (2011).
Tao, R. et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nature medicine 13, 1299–1307 (2007).
Wells, A. D. New insights into the molecular basis of T cell anergy: anergy factors, avoidance sensors and epigenetic imprinting. Journal of immunology 182, 7331–7341 (2009).
Mason, K. L. et al. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infection and immunity 80, 3371–3380 (2012).
Gillilland, M. G., 3rd et al. Ecological Succession of Bacterial Communities during Conventionalization of Germ-Free Mice. Appl Environ Microbiol 78, 2359–2366 (2012).
Mason, K. L. et al. Interplay between the gastric bacterial microbiota and Candida albicans during postantibiotic recolonization and gastritis. Infect Immun 80, 150–158 (2012).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541 (2009).
vegan: Community Ecology Package. R package version 1.17-3 v. 1.17-3 (2010).
Cole, J. R. et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic acids research 37, D141–145 (2009).
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology 73, 5261–5267 (2007).
gplots: Various R programming tools for plotting data. v. R package version 2.10.1 (2011).
Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62, 142–160 (2007).
Ter Braak, C. J. F. Canonical Correspondence Analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67, 1167–1179 (1986).
Russell, C. & Lay, K. M. Natural history of Candida species and yeasts in the oral cavities of infants. Arch Oral Biol 18, 957–962 (1973).
Scupham, A. J. et al. Abundant and diverse fungal microbiota in the murine intestine. Appl Environ Microbiol 72, 793–801 (2006).
Iliev, I. D. et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336, 1314–1317 (2012).
Morales, D. K. & Hogan, D. A. Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog 6, e1000886 (2010).
Kennedy, M. J. & Volz, P. A. Effect of various antibiotics on gastrointestinal colonization and dissemination by Candida albicans. Sabouraudia 23, 265–273 (1985).
Jacobus, N. V., Tally, F. P., Barza, M. & Gorbach, S. L. Susceptibility of anaerobic bacteria to cefoperazone and other beta-lactam antibiotics. Clin Ther 3, 34–38 (1980).
Reeves, A. E., Koenigsknecht, M. J., Bergin, I. L. & Young, V. B. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun 80, 3786–3794 (2012).
Brown, A. J. et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem 278, 11312–11319 (2003).
Gomez-Hurtado, I. et al. Gut microbiota dysbiosis is associated with inflammation and bacterial translocation in mice with CCl4-induced fibrosis. PLoS One 6, e23037 (2011).
Sina, C. et al. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J Immunol 183, 7514–7522 (2009).
Philpott, D. J. & Girardin, S. E. Nod-like receptors: sentinels at host membranes. Curr Opin Immunol 22, 428–434 (2010).
Ratner, A. J., Aguilar, J. L., Shchepetov, M., Lysenko, E. S. & Weiser, J. N. Nod1 mediates cytoplasmic sensing of combinations of extracellular bacteria. Cell Microbiol 9, 1343–1351 (2007).
Cosi, C. et al. G-protein coupled receptor 35 (GPR35) activation and inflammatory pain: Studies on the antinociceptive effects of kynurenic acid and zaprinast. Neuropharmacology 60, 1227–1231 (2011).
Fallarini, S., Magliulo, L., Paoletti, T., de Lalla, C. & Lombardi, G. Expression of functional GPR35 in human iNKT cells. Biochem Biophys Res Commun 398, 420–425 (2010).
Carvalho, F. A., Aitken, J. D., Vijay-Kumar, M. & Gewirtz, A. T. Toll-like receptor-gut microbiota interactions: perturb at your own risk!. Annu Rev Physiol 74, 177–198 (2012).
Cinova, J. et al. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: study in germ-free rats. PLoS One 6, e16169 (2011).
Ouellette, A. J. Paneth cells and innate mucosal immunity. Curr Opin Gastroenterol 26, 547–553 (2010).
Vaishnava, S. et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011).
Erb-Downward, J. R. & Noverr, M. C. Characterization of prostaglandin E2 production by Candida albicans. Infect Immun 75, 3498–3505 (2007).
Haas-Stapleton, E. J. et al. Candida albicans modulates host defense by biosynthesizing the pro-resolving mediator resolvin E1. PloS one 2, e1316 (2007).
Noverr, M. C., Phare, S. M., Toews, G. B., Coffey, M. J. & Huffnagle, G. B. Pathogenic yeasts Cryptococcus neoformans and Candida albicans produce immunomodulatory prostaglandins. Infect Immun 69, 2957–2963 (2001).
Acknowledgements
We would like to acknowledge Roderick McDonald, Katy Higdon and Kelly Mason for their technical support of this project. This work was supported by NIAID grants R21-AI087869 (GBH) and U19AI090871(GBH) and NIH NIDDK grant P30DK034933 (GBH).
Author information
Authors and Affiliations
Contributions
Designed project: J.E.D., K.M. and G.H. Performed experiments: K.M., N.F. and J.E.D. Analyzed Data: J.E.D., G.H., J.E.D. and R.M. Generated Figures: J.E.D., G.H. and R.M. Wrote the manuscript: J.E.D. and G.H. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary
Rights and permissions
This work is licensed under a Creative Commons Attribution 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/
About this article
Cite this article
Erb Downward, J., Falkowski, N., Mason, K. et al. Modulation of Post-Antibiotic Bacterial Community Reassembly and Host Response by Candida albicans. Sci Rep 3, 2191 (2013). https://doi.org/10.1038/srep02191
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02191
This article is cited by
-
Characterization of the intestinal fungal microbiome in patients with hepatocellular carcinoma
Journal of Translational Medicine (2023)
-
The interactions of Candida albicans with gut bacteria: a new strategy to prevent and treat invasive intestinal candidiasis
Gut Pathogens (2023)
-
Maintenance of gut microbiome stability for optimum intestinal health in pigs – a review
Journal of Animal Science and Biotechnology (2022)
-
Performance of distinct microbial based solutions in a Campylobacter infection challenge model in poultry
Animal Microbiome (2022)
-
“Molding” immunity—modulation of mucosal and systemic immunity by the intestinal mycobiome in health and disease
Mucosal Immunology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.