Abstract
Phagocytosis is crucial for triggering host defenses against invading pathogens in animals. However, the receptors on phagocyte surface required for phagocytosis of virus have not been extensively explored. This study demonstrated that white spot syndrome virus (WSSV), a major pathogen of shrimp, could be engulfed but not digested by Drosophila S2 cells, indicating that the virus was not recognized and taken up by a pathway that was silent and would not activate the phagosome maturation and digestion pathway. The results showed that the activation of receptors on S2 cell surface by lipopolysaccharide or peptidoglycan resulted in the phagocytosis of S2 cells against WSSV virions. Gene expression profiles revealed that the dally-mediated Wnt signaling pathway was involved in S2 phagocytosis. Further data showed that the Wnt signaling pathway played an essential role in phagocytosis. Therefore, our study contributed novel insight into the molecular mechanism of phagocytosis in animals.
Similar content being viewed by others
Introduction
Phagocytosis, a highly conserved process, is crucial for the immune responses of animals and it allows for rapid engulfment of pathogens and apoptotic cells by specialized phagocytes1,2,3,4,5. During phagocytosis, outer particles are recognized by binding to the surface of phagocytes, engulfed by cytoskeleton rearrangement to form the phagosome and are then digested in a mature phagosome4. Phagocytes discriminate among diverse particles through the array of receptors expressed on their surfaces5. Receptors bound with particles trigger an orderly progression of cellular changes leading to rearrangement of the actin cytoskeleton and membrane remodeling. Central to these processes is the generation of a phagosome6. Once formed, the phagosome undergoes maturation by fission and limited fusion events with endosomes, lysosomes, or both to generate the mature phagolysosome7,8. Pathogens inside the phagolysosome are destroyed by low pH, hydrolysis and radicals. However, the receptors on phagocyte surface required for phagocytosis of virus have not been extensively explored.
Generally, it is believed that the phagocytosis process is initiated by the ligation of cell surface receptors that either directly bind to the particles or the opsonins that are deposited on the particle's surface. To date, the Fc receptor of immunoglobulin (Ig) on phagocyte surface greatly contributes to our knowledge about receptors and their pathways responsible for phagocytosis9,10,11,12. The in vitro studies have confirmed the roles of the Fc receptor in triggering effector responses such as macrophage phagocytosis13,14. The receptor is relocated to phagolysosomes during phagocytosis of IgG opsonized bacteria15. In invertebrates, the pattern recognition receptors play essential roles in the antibacterial immunity. The activated pattern recognition receptors trigger Imd and Toll pathways in fighting bacteria and fungi infections16,17,18,19. The receptor, peptidoglycan (PG) recognition protein LC (PGRP-LC), induces phagocytosis of Gram-negative bacteria via activation of the Imd signaling pathway16,17,20,21. The Toll pathway induced by PGRP-SA (SD) and Gram-negative-binding protein (GNBP1 or GNBP3) leads to the activation of the phagocytosis of Gram-positive bacteria and fungi16,17,22. As reported, lipopolysaccharide (LPS) from Gram-negative bacteria can activate the Imd signaling pathway. To date, however, no IgG-like receptor has ever been found on phagocytes from invertebrates due to the lack of the acquired immunity in invertebrates. Therefore, it is believed that there are some specific receptors and their corresponding signaling pathways in phagocytes of invertebrates that are comparable to the Ig receptors on invertebrate phagocytes. But this issue has not yet been addressed.
We have found that white spot syndrome virus (WSSV), a major pathogen of shrimp, could be engulfed by Drosophila melanogaster Schneider line 2 cells (S2 cells), a phagocytic cell line. However, the WSSV virions were not digested by S2 cells, leading to the consideration that the virus was recognized and engulfed by phagocytes, but phagosome maturation was not occurring in S2 cells against WSSV virions. The results further revealed that the dally-receptor-mediated Wnt signaling pathway played an essential role in phagocytosis in Drosophila.
Results
Requirement of specific receptors on phagocyte surface to trigger phagocytosis
In order to evaluate the infectivity of WSSV in Drosophila, S2 cells were inoculated with WSSV. To observe the distribution of the virions within the cells, the WSSV-inoculated cells were examined using transmission electron microscopy (TEM). The results showed that the WSSV virions were engulfed and enveloped by vesicles for 5 days (Fig. 1A). This phenomenon indicated that WSSV, as a non-pathogenic virus of Drosophila, could not be digested by S2 cells which have strong phagocytic activity. Quantitative real-time polymerase chain reaction (RT-PCR) data showed that the viral copies did not change in S2 cells at 1–5 days after WSSV inoculation (Fig. S1), suggesting that the WSSV virions were engulfed but did not replicate in S2 cells. These findings suggested that WSSV could be recognized and engulfed by S2 cells but not digested/degraded and this virus was not infectious since it could not replicate inside S2 cells and trigger its release from these S2 cells to further propagate and infect other cells.
The phagocytosis of microbes by Drosophila S2 cells.
(A) The white spot syndrome virus (WSSV) virions in S2 cells. The S2 cells were inoculated with WSSV. At 5 d post-inoculation, the S2 cells were examined using transmission electron microscopy (TEM) (right). The box indicated the enlarged WSSV virions in a vesicle. The S2 cells without WSSV were used as a control. Scale bar: control, 2 μm; WSSV, 1 μm. (B) The phagocytosis of microbes by S2 cells using TEM. Pathogenic and non-pathogenic bacteria (Escherichia coli and Shewanella xiamenensis) as well as pathogenic and non-pathogenic viruses (DCV and WSSV) were inactivated and then inoculated into S2 cells for the phagocytosis assays. Non-inactivated WSSV [WSSV (live)] was included in the inoculation as a control. The S2 cells were examined at different time points after inoculation using TEM. The boxes showed enlarged images. The numbers indicated the time after inoculation. Scale bar, 1 μm. (C) S2 phagocytosis of microbes under confocal microscopy (100×). Inactivated pathogenic and non-pathogenic bacteria or viruses as well as the non-inactivated WSSV were labeled with fluorescein isothiocyanate (FITC). The S2 cells were then inoculated with the FITC-labeled microbes. The lane headings showed the microbes used. At 15 min and 24 hr after inoculation, the S2 cells were fixed and stained with rhodamine phalloidin to label the actin (red) and with 4′,6-diamidino-2-phenylindole to label the DNA (blue), followed by examination using confocal microscopy. The FITC-labeled microbes (green) were indicated with arrows.
Pathogenic and non-pathogenic bacteria (Escherichia coli and Shewanella xiamenensis, respectively) as well as pathogenic and non-pathogenic viruses (Drosophila C virus [DCV] and WSSV, respectively) were used to evaluate whether pathogenic and non-pathogenic bacteria and viruses could elicit the same innate immune response and phagocytic response. The inactivated bacteria, inactivated viruses and non-inactivated WSSV [WSSV (live)] were inoculated into S2 cells for the phagocytosis assays. The TEM results showed that the inactivated E. coli and S. xiamenensis were engulfed in S2 cells at 1 hr after inoculation (Fig. 1B). At day 1 after inoculation, they were completely digested in the S2 cells (Fig. 1B). Because E. coli is a pathogenic bacterium of D. melanogaster, lipopolysaccharide (LPS) located on the outer membrane of E. coli can activate the Imd pathway via Drosophila peptidoglycan recognition protein LC (PGRP-LC), leading to the activation of phagocytes. Peptidoglycan (PG), which is distributed on the outer membrane of S. xiamenensis, a non-pathogenic bacterium of D. melanogaster, can initiate the Toll pathway via PGRP-SA (SD) and Gram-negative bacteria binding protein 1 (GNBP-1), resulting in the activation of S2 phagocytosis. These data suggested that both the pathogenic and the non-pathogenic bacteria could be digested by S2 cells. It was shown that both the inactivated pathogenic virus (DCV) and non-pathogenic virus (WSSV) were engulfed and subsequently digested in S2 cells (Fig. 1B), yielding the same results as those of the pathogenic and non-pathogenic bacteria. However, the non-inactivated WSSV [WSSV (live)] was engulfed but not digested by S2 cells even at 5 days after the WSSV incubation (Fig. 1B).
Phagocytosis of these bacteria and viruses was examined by confocal microscopy to further confirm the TEM results. The results demonstrated that the inactivated pathogenic or non-pathogenic bacteria and viruses were observed in S2 cells at 15 min after inoculation and then digested by S2 cells at 24 hr after inoculation (Fig. 1C). However, the non-activated WSSV [WSSV (live)] could not be digested by S2 cells at all examined time points (Fig. 1C). The results were the same as those of TEM. This demonstrated that the virus itself was capable of subverting the host cell immune system to prevent its targeting to phagosome-lysosome degradation via maturation of the phagosome.
Initiation of phagocytosis by receptors recognizing LPS or PG
The time course results showed that the inactivated E. coli was engulfed by S2 cells at 15–60 min after inoculation and then digested in 24 hr after inoculation (Fig. 2A). On the other hand, the non-inactivated WSSV virions [WSSV (live)] were also engulfed but not digested by S2 cells and the virions could not replicate and escape S2 cells to infect other cells, thereby being non-infectious (Fig. 2A). To activate the immune system by stimulating the Imd and Toll pathways using LPS and PG, the S2 cells were treated with LPS or PG for 1 hr followed by WSSV inoculation. The results showed that the LPS- or PG-activated S2 cells could engulf and subsequently digest the WSSV virions (Fig. 2A), indicating that the activation of the Imd or Toll pathways could promote the degradation of the live WSSV presumably by restoring phagosome maturation.
LPS and PG -induced S2 phagocytosis of WSSV.
(A) The phagocytic process of fluorescein isothiocyanate (FITC)-labeled microbes under confocal microscopy (400×). The FITC-labeled inactivated Escherichia coli or non-inactivated WSSV virions [WSSV (live)] were inoculated into S2 cells. LPS or PG was included in the inoculation to activate the receptors recognizing LPS or PG. The cells were fixed at different time points after inoculation. The cells were then stained with rhodamine phalloidin to label the actin and with DAPI to label the DNA. The numbers indicated the examined time points. (B) Phagocytosis of pHrodo-labeled microbes under confocal microscopy. The pHrodo-labeled inactivated E. coli, inactivated WSSV, or WSSV (live) were inoculated into S2 cells. LPS or PG was simultaneously inoculated. At 1 hr after inoculation, the cells were examined using confocal microscopy (up, 100×). The red fluorescence inside the S2 cells represented the phagolysosome acidification. The images were enlarged (down, 400×). (C) S2 phagocytosis percentages of the pHrodo-labeled microbes. The number of S2 cells phagocytosing the pHrodo-labeled microbes was quantified using flow cytometry at 1 hr after microbial inoculation. Non-treated S2 cells were used as a control. Statistically significant differences between treatments were indicated with asterisks (** P < 0.01).
During maturation, the phagosome fuses with the lysosomes resulting in its acidification which allows for the digestion of engulfed particles. A pH-sensitive dye, pHrodo can be used to label bacteria and monitor their engulfment in a fully mature phagosome simply by examining whether these pHrodo-labeled bacteria fluoresce when found in this acidified environment. To investigate the phagocytosis of WSSV by LPS- or PG-activated S2 cells, non-activated (live) WSSV virions were labeled with pHrodo and examined by confocal microscopy. The results indicated that inactivated E. coli and WSSV were engulfed by S2 cells into an acidified mature phagosome at one hour after inoculation, whereas the non-inactivated WSSV virions could not induce the phagolysosome acidification (Fig. 2B). When the S2 cells were pretreated with LPS or PG activating the Imd or Toll pathway, respectively, live WSSV was taken up by S2 cells into a fully mature phagosome (Fig. 2B) at one hour after inoculation, showing that the engulfed non-inactivated WSSV virions were digested/degraded into fully mature acidified phagosome.
To evaluate the effects of LPS or PG receptor activation on phagocytosis, the phagocytic percentage of LPS- or PG-activated S2 cells against the pHrodo-labeled WSSV was characterized. The data showed that the phagocytic percentage of LPS- or PG-activated S2 cells was significantly increased (P < 0.01) compared with the control at 1 hr after WSSV inoculation (Fig. 2C). S2 cells inoculated with the pHrodo-labeled inactivated WSSV or inactivated E. coli yielded similar phagocytosis results (Fig. 2C). However, the percentage of phagocytic S2 cells inoculated with the non-inactivated WSSV virions (live WSSV) only was very low, which was similar to that of the control. The results suggested that activation of the LPS- or PG-induced Imd and Toll pathways favored the uptake of live WSSV through a phagocytic pathway that allowed for the uptake and digestion of this virus into a fully matured phagosome.
Identification of the signaling pathways required for phagocytosis
To assess effects of the pathogenic DCV and non-pathogenic WSSV on the gene expressions of S2 cells, the gene expression profiles of S2 cells treated with DCV and WSSV were characterized using the GeneChip Drosophila Genome 2.0 Array. The array analysis showed that the gene expression profiles of S2 cells treated with inactivated or non-inactivated DCV or WSSV differed largely from those of the control (non-treated S2 cells) (Fig. 3a). Compared with the control, 802, 737, 524 and 877 genes were altered by at least two fold for inactivated DCV, non-inactivated DCV [DCV (live)], inactivated WSSV and non-inactivated WSSV [WSSV (live)], respectively (Fig. 3a). The results showed that 626 genes (250 up-regulated genes and 376 down-regulated genes) in S2 cells treated with inactivated or non-inactivated DCV had the same gene expression patterns, indicating that infection and phagocytosis of the pathogenic virus might share many same or similar pathways. However, the gene expression profiles of S2 cells treated with inactivated or non-inactivated WSSV differed largely (Fig. 3a), suggesting that the live non-pathogenic virus activated different pathways than the non-pathogenic inactivated virus. The results were consistent with the TEM and confocal microscopy findings.
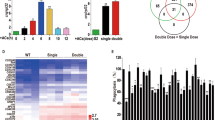
Receptors and signaling pathways required for phagocytosis of virus.
(a) The gene expression profiles of Drosophila genes in response to virus. The gene expression profiles were conducted with a DNA microarray using S2 cells treated with inactivated Drosophila C virus (DCV) or white spot syndrome virus (WSSV) and non-inactivated DCV [DCV (live)] or WSSV [WSSV (live)] (up). Non-treated S2 cells were used as a control. Red indicated the up-regulation and green showed the down-regulation. The differentially expressed genes were evaluated (down). The numbers represented the genes up-regulated or down-regulated (more than twofold) compared with the control. (b) The biological processes mediated by the differentially expressed genes. (c) The gene network of the dally mediated pathways. The green and blue lines indicated the GenMAPP and KEGG databases, respectively. The numbers indicated the times that two genes occurred in a pathway. (d) The model of the dally mediated pathways in phagocytosis. The dally receptor was distributed on the cell membrane. The solid lines showed the known interactions or pathways. The dashed lines indicated the unknown interactions or pathways.
In order to identify the pathways being activated in S2 cells in response to inactivated and non-inactivated DCV and inactivated and non-inactivated WSSV, the DNA microarray data were further analyzed. The results showed that 120 genes shared the same expression profiles (up-regulation or down-regulation) in three treatments [inactivated DCV, DCV (live) and inactivated WSSV] but not in WSSV(live) treatment, suggesting that these genes were required for phagosome maturation and digestion of inactivated and live DCVs and inactivated WSSVs. Among these genes, 53 genes were up-regulated and 67 genes were down-regulated. The analysis showed that the 120 genes participated in diverse biological processes including defense responses, signal transduction, cytoskeleton regulation, vesicle transport, apoptosis and proteolysis (Fig. 3b).
These down-regulated and up-regulated genes were further analyzed to investigate the cell receptors and receptor-mediated pathways involved in phagocytosis. Gene ontology and KEGG analysis revealed that the dally (division abnormally delayed) receptor and its corresponding Wnt pathway was involved in phagocytosis (Fig. 3C). The results showed that dally receptor, located on the cell surface, could interact with eight proteins including amphiphysin (Amph), glutactin (Glt), inscuteable (insc), pericardin (prc), heartless (htl), matrix metalloproteinase 1 (Mmp1), frizzled 2 (fz2) and disheveled associated activator of morphogenesis (DAAM) (Fig. 3c). Among the eight proteins, fz2 and DAAM were the components of the Wnt signaling pathway. We found that DAAM interacted with myosin 31DF (Myo31DF), a cytoskeletal protein that demonstrated actin binding and motor activities. These data suggested that the Wnt pathway may be involved in phagosome maturation (Fig. 3d). The analysis showed that Amph (required in endocytosis of synaptic vesicles and vesicle fission), Glt (acidic sulphated glycoprotein of basement membrane) and insc (a cytoskeletal adaptor) could interact with the dally receptor (Fig. 3c). In this context, the interaction between the dally receptor and Amph, Glt, or insc led to the suggestion that the dally receptor played a key role in phagosome maturation through regulating cytoskeletal organization (Fig. 3d). The findings indicated that prc, htl and Mmp1 might be related to phagosome maturation, but the corresponding pathways mediated by the three proteins were unknown. Taken together, the results suggested that the dally receptors on the cell surface might mediate Wnt signaling pathway that was involved in phagosome maturation.
Roles of the dally-mediated Wnt pathway in phagocytosis
In order to evaluate the roles of the dally-mediated pathways in phagocytosis, we examined the expression profiles of eight genes (Amph, Glt, insc, prc, htl, Mmp1, fz2 and DAAM) encoding proteins that interacted with dally, as well as dally and Myo31DF, using S2 cells in response to LPS or PG stimulation. The quantitative RT-PCR results indicated that the expressions of dally, Amph, Glt, prc, htl, fz2, DAAM and Myo31DF were very significantly up-regulated (P < 0.01) compared to that of the control gene rp49 after LPS or PG stimulation (Fig. 4A). The results showed that the dally-mediated pathways (Fig. 3d) were involved in phagosome maturation. However, the expression profiles of insc and Mmp1 did not change after LPS or PG stimulation (Fig. 4A). Of the dally-mediated pathways (Fig. 3d), fz2 and DAAM belong to the Wnt pathway. To characterize the role of Wnt pathway in phagosome maturation, we detected the gene expression profiles of the Wnt pathway in response to LPS or PG stimulation. The results indicated that dally, wnt2, fz2, dsh, DAAM, sgg and arm were very significantly up-regulated (P < 0.01) after LPS or PG stimulation (Fig. 4A and B). The data suggested that the Wnt pathway may take great effects on phagosome maturation.
Roles of the dally-mediated pathways in phagocytosis.
(A) The expression profiles of genes belonging to the dally-mediated pathways in S2 cells in response to LPS or PG challenge. The rp49 gene was used as a control. Statistically significant differences between treatments were indicated with asterisks (** P < 0.01). Control, non-treated S2 cells. (B) The expression profiles of other genes belonging to the Wnt pathways in S2 cells in response to LPS or PG challenge. (C) Effects of dally gene silencing on gene expressions of the Wnt pathway. To evaluate the involvement of dally in phagocytosis, expression of the dally gene in S2 cells was silenced using sequence-specific siRNA. The expression profiles of the genes related to the dally-mediated pathways were then examined using quantitative RT-PCR. The rp49 gene was used as a control. Statistically significant differences between treatments were indicated with asterisks (*P < 0.01 or **P < 0.01). Control, non-treated S2 cells. (D) Effects of dally gene silencing on expressions of genes belonging to the dally-mediated pathways. (E) Percentages of S2 phagocytosis against the pHrodo-labeled WSSV virions. The number of S2 cells phagocytosing the pHrodo-labeled WSSV virions was quantified using flow cytometry at 1 hr after virus inoculation. The inactivated WSSV (WSSV) and non-inactivated WSSV [WSSV (live)] were included in the experiments. Non-treated S2 cell was used as a control. Statistically significant differences between treatments were indicated with asterisks (** P < 0.01). (F) The model of the dally-mediated pathways involved in phagocytosis. The Wnt pathway was shown in the box.
To further evaluate the role of dally in phagosome maturation, the dally gene was silenced using sequence-specific siRNA in S2 cells. We investigated the expression levels of wnt2, fz2, dsh, DAAM, sgg and arm to reveal the effects of dally gene silencing on the expression of genes belonging to the Wnt pathway. The results showed that dsh and arm were very significantly down-regulated (P < 0.01) whereas sgg and wnt2 were very significantly up-regulated (P < 0.01) when dally was knocked down (Fig. 4C). The silencing of dally gene expression led to up-regulation or down-regulation of seven genes (Amph, Glt, prc, htl, fz2, DAAM and Myo31DF) that were involved in dally-mediated pathways (Fig. 4D). However, insc and Mmp1 gene expressions were not affected largely. The data were consistent with the quantitative RT-PCR results. We found that Amph, prc, fz2 and Myo31DF were significantly up-regulated (P < 0.05) whereas Glt, htl and DAAM were significantly down-regulated (P < 0.05) when dally was knocked down (Fig. 4D). The results showed that the phagocytic percentage of pHrodo-labeled inactivated WSSV in dally-siRNA-treated S2 cells was significantly decreased (P < 0.01) compared with that of the control at 1 hr after inoculation (Fig. 4E). LPS and PG could induce the uptake and digestion of live WSSV into a fully matured phagosome (Fig. 4E). The silencing of dally expression significantly inhibited the phagosome maturation process. These findings suggested that the dally-mediated Wnt pathway might be essential for phagocytosis (Fig. 4F).
Discussion
Widespread evidence indicates that phagocytosis, an actin-dependent process involving the internalization of bacteria and viruses, represents an essential branch of the immune system of organisms2,23. The phagocytic process begins with particle recognition and binding with receptors on host cell surface, leading to the engulfment of particles into cell and subsequent formation of phagosomes to initiate the digestion of microbes. Phagocyte-microbe contact is accompanied by intracellular signals that trigger diverse cellular processes such as cytoskeletal rearrangement, alterations in membrane trafficking and activation of microbial killing mechanisms24. It is believed that the engulfment of particles into cell represents the initiation of phagocysotis. In this study, the results showed that the non-pathogenic WSSV could be engulfed by phagocytes (S2 cells), but the WSSV virions were not digested in S2 cells, suggesting that the phagosome maturation might not occur. The findings indicated that the engulfed particles could be digested by phagocytes when the phagosome maturation was induced. Now that the engulfment of particles by phagocytes does not represent triggering the digestion of engulfed particles, the strategy used for the evaluation of phagocytosis with fluorescently labeled latex beads25,26,27 needs to be improved. The fluorescently labeled latex beads may be used to assess the ability of phagocyte engulfment only. If they are used for examining the phagocytic activity of phagocytes, the beads can be coated with LPS or PG to induce the phagosome maturation leading to digestion/degradation of particles.
Our study demonstrated that the non-pathogenic virus WSSV would not be completely digested by phagocytes, although the WSSV virions were engulfed into S2 cells. Recently this similar phenomenon was observed in Ehrlichia chaffeensis, a member of the order Rickettsiales. It was found that Drosophila S2 cells were permissive for the growth of E. chaffeensis and the cells could completely digested E. chaffeensis after LPS stimulation23. It is well documented that the recognition of bacterial and fungal pathogens by receptors on cell surface leads to activations of the Toll and Imd pathways in invertebrates16,17,21. Lipopolysaccharide (LPS), a major component of the outer membranes of gram-negative bacteria, is one of the most powerful activators of macrophages via the Imd pathway, while peptidoglycan (PG), a major component of gram-positive bacterial cell walls, can activate macrophages via the Toll pathway28,29. Recent studies show that some microbes can be engulfed but not digested by the phagocytes of mammals and insects20,30. These findings suggest that some receptors, except for the known receptors, are essential for phagocytosing intracellular pathogens efficiently by host phagocytes. The present and previous studies show that activation of the LPS- or PG-induced Imd and Toll pathways favor the uptake and digestion of microbes into a fully matured phagosome31,32. Up to date, the well-documented receptor for phagocytosis is Fc receptor in vertebrates32. In this context, there must be some unrevealed receptors on invertebrate phagocyte surface to trigger the phagocytosis response, which may be comparable to the Fc receptor. In this study, based on the genome-wide microarray analysis of Drosophila, the results showed that the dally-mediated Wnt signaling pathway played important roles in phagocytosis. As reported, the Wnt pathway is highly conserved between flies and humans despite the evolutionary distance between the species33. The Wnt family of signaling proteins participates in multiple developmental events during embryogenesis and Wnt signals are pleiotropic, with effects that include mitogenic stimulation, cell fate specification and differentiation34,35. Recently, it is found that the Wnt-induced endocytosis can initiate synapse formation and endosomes may constitute a crucial signaling organelle36,37,38. In our study, the results indicated that the dally-mediated Wnt pathway took effects on phagocytosis. This finding contributed a novel aspect of Wnt pathway in immune responses of animals and the dally-mediated pathways in phagocytosis merited to be further investigated.
Methods
Shrimp culture, WSSV purification and WSSV inoculation in drosophila S2 cells
Cultures of Marsupenaeus japonicus shrimp, approximately 10 g and 10–12 cm each, were performed keeping in groups of 20 individuals in 80 L aquariums at 20°C. Then the virus-free shrimp were infected with WSSV. The WSSV-infected shrimp were collected and homogenized for virus purification according to the previous methods39. For quantitative analysis of viral DNA, the real-time PCR was conducted as described before40. The WSSV virions were UV-inactivated by exposure to a total of 12,000 mJ UV light (5 × 3 min). Subsequently the WSSV virions (1 × 107 copies/mL) or inactivated WSSV virions (1 × 107 copies/mL) were inoculated in Drosophila S2 cells (1 × 106 cells/mL) for culture at 28°C, respectively. At different time, the S2 cells were collected and subjected to transmission electron microscopy, confocal microscopy, DNA microarray and quantitative real-time PCR analyses.
Maintenance of drosophila S2 cell Line and drosophila C virus (DCV) purification and infection
Drosophila S2 cells were cultivated at 28°C in Schneider's Drosophila medium (Ivitrogen, USA) supplemented with 10% fetal bovine serum (Gibco, USA). DCV was inoculated in S2 cells at a multiplicity of infection (MOI) of 1 for 5 days and collected for purification as described before1. DCV at a multiplicity of infection (MOI) of 1 was UV-inactivated by exposure to a total of 12,000 mJ UV light (5 × 3 min) as inactivated virus. Then S2 cells were inoculated with UV-inactivated DCV. At different time, the S2 cells were collected and subjected to transmission electron microscopy, confocal microscopy, DNA microarray and quantitative real-time PCR analyses.
Culture of bacteria and bacterial inoculation in S2 cells
Escherichia coli DH5α strain was grown in Luria–Bertani (LB) medium at 37°C for 24 hr and collected by centrifugation at 10,000 × g for 10 min. Shewanella xiamenensis strain S4T was obtained from a sediment sample that was collected from a coastal area of Xiamen, China41. The strain was grown in LB medium at 30°C for 24 hr and collected by centrifugation at 10,000 × g for 10 min. E. coli and S. xiamenensis were inactivated at 80°C for 1 hr, respectively. Then S2 cells were inoculate with inactivated bacteria (heat-inactivated E. coli or S. xiamenensis) at a density of 5 × 106/mL and incubated for 1 hr at 28°C. At different time, the S2 cells were collected for analysis.
Activation of S2 phagocytosis with lipopolysaccharide (LPS) or peptidoglycan (PG)
To determine whether the activation of receptors recognizing LPS or PG on S2 cell surface could initiate the phagocytosis of WSSV, the S2 cells were plated at a concentration of 1 × 106 cells well in six-well plates and were allowed to adhere for at least 30 min. The LPS or PG (Sigma, USA) was sonicated for 1 hr and then added to S2 cells at a concentration of 10 μg per ml. After incubation with LPS or PG for 1 hr, the S2 cells were inoculated with the purified WSSV virions for 15 min-1 hour.
Transmission electron microscopy assay
The S2 cells were pelleted and fixed in the fixative containing 2% paraformaldehyde and 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 18 hr at room temperature. Each sample was washed three times with 0.1 M sodium cacodylate buffer at room temperature. Then the sample was postfixed with 2% osmium tetroxide in 0.1 M sodium cacodylate buffer with constant rotation for 1 hr, followed by washes three times using 0.1 M sodium cacodylate buffer at room temperature. The sample was stained with 2% uranyl acetate in 0.2 M sodium acetate buffer (pH 5.2) for 1 hr at room temperature and subsequently washed three times with 0.2 M sodium acetate buffer at room temperature. The sample was dehydrated in an ascending acetone series (50, 60, 70, 80, 90, 95 and 100%) and then in 100% propylene oxide for 10 min with constant rotation at room temperature. After infiltration of sample with EMBED 812/Araldite 502 resin at room temperature, sections were prepared in a Reichert Ultracut OMU3 microtome (Leica, Germany) at 100 nm thickness, followed by staining with 2% uranyl acetate/70% methanol. The images were collected on a Hitachi 7650 transmission electron microscope (Hitachi, Japan) operating at 70 kV.
Confocal microscopy analysis
Drosophila S2 cells were plated in 12-well plastic tissue culture plate in 1 ml of Schneider's medium with 10% FBS. FITC-labeled (Invitrogen, USA) microbes were added to each well containing S2 cells at a density of 5 × 106, followed by incubation for 1 hr at 28°C. After washes with PBS, the S2 cells were transferred to Poly-Prep glass slides (Sigma, USA) and fixed. Then the S2 cells were incubated with rhodamine phalloidin (Invitrogen, USA) or DAPI (Sigma, USA) to label actin or nucleus DNA, respectively. Fluorescent images were taken with a Zeiss Laser scanning systems LSM 510 Meta (Carl Zeiss, Germany) and processed using Zeiss LSM Image Examiner Version software.
Detection of phagolysosome acidification with pHrodo labeling
The pHrodo dye, a novel fluorogenic dye that dramatically increased the fluorescence as the pH of its surroundings became more acidic42, was used to label E. coli or WSSV using the pHrodo phagocytosis particle labeling kit (Invitrogen, USA) according to the manufacturer's manual. The unique pHrodo-based system could measure the phagocytic activity based on acidification of particles as they were ingested. The S2 cells (1 × 106) were inoculated with the pHrodo-labeled inactivated E. coli (5 × 106/mL), inactivated WSSV virions (1 × 107 copies/mL) or non-inactivated WSSV virions (1 × 107 copies/mL), respectively. After incubation at 28°C for 1 hr, the S2 cells were collected and immediately analyzed with confocal microscopy. To activate the LPS- or PG- mediated receptors of phagocytosis, the pHrodo-labeled inactivated E. coli, inactivated WSSV or non-inactivated WSSV were co-inoculated with LPS or PG into S2 cells. The percentages of S2 cells phagocytosed pHrodo-labeled microbes were calculated. The differences between different treatments were statistically analyzed by t-test assuming unequal variances. A P-value < 0.05 or < 0.01 was considered to be significant.
Analyses of gene expressions with oligonucleotide microarray
S2 cells (1 × 106) were inoculated with DCV at a multiplicity of infection (MOI) of 1 for 1 hr. Inactivated DCV was treated in the same way. The WSSV virions (1 × 107 copies/mL) or inactivated WSSV virions (1 × 107 copies/mL) were inoculated into S2 cells (1 × 106 cells/mL) for 1 hr. Then total RNA was extracted from treated S2 cells by using Trizol reagent (Invitrogen, USA) according to the manufacturer's instructions. The total RNA samples were then analyzed by CapitalBio Corp for Chip (Affymetrix) assay. Gene expression analysis was performed by using the Affymetrix (Santa Clara, CA, USA) Drosophila GeneChip, using the laboratory methods in the Affymetrix GeneChip expression manual. Gene expression analysis was performed using multiple arrays and multiple independent mRNA samples for each treatment. Microarray data were analyzed by using Bio MAS (molecule annotation system) 3.0 software (CapitalBio Corporation, Beijing, China). Using the criterion of cutoff limitation as a fold change ≥2 or ≤0.5 and q-value ≤5%, differential expression genes were screened and clustered.
Biological pathway analysis
Through array analysis, the commonly altered genes were screened from DCV and WSSV treatments. The selected genes were further analyzed in the context of Gene Ontology (GO) biological process and Kyoto Encyclopaedia of Genes and Genomes (KEGG) biological pathway. Then the results of signaling pathway and protein interaction about these genes were analyzed by MAS 3.0 software. To reveal the functions of predicted target genes, we used the ontology classification of genes based on gene annotation and summary information available through DAVID (Database for Annotation, Visualization and Integrated Discovery).
RNAi assays in S2 cells
To silence the expression of dally gene, the dally-specific siRNA (5′-GCGAAAC GAAAUUGUGCUATT-3′) was prepared using a Takara T7 in vitro transcription kit (Takara, Japan) according to the manufacturer's protocol. The siRNAs were quantified by Nanodrop 2000 Spectrophotometer (Thermo, USA) and stored at −80°C for use. RNAi in S2 cells was performed with Lipofectamine RNAi MAX Transfection Reagent (Invitrogen, USA) according to the manufacturer's instructions.
Quantitative real-time PCR
Total RNAs were extracted from S2 cells (1 × 106) using Trizol reagent (Invitrogen, USA) according to the manufacturer's instructions. Then the total RNA (5 μg) was reversely transcribed to synthesize cDNA in a volume of 20 μL with reverse transcriptase M-MLV (Takara, Japan). Quantitative real-time PCR was performed using gene-specific primers and TaqMan probe (Table S2). Relative expression levels of interest genes were calculated as previously described43, using rp49 as an internal control in S2 cells. Real-time PCR was carried out in a final volume of 20 μl, which contained 10 μl Premix Ex Taq (TaKaRa, Japan), 1 μl diluted cDNA template, 7.2 μl distilled water, 0.3 μl of each TaqMan probe and 0.4 μl of each primer. PCR conditions were as follows, 95°C for 30 seconds, followed by 50 cycles of 95°C for 5 seconds and 60°C for 30 seconds. Resultant data were analyzed using the iQTM5 software.
WSSV quantitative analysis by TaqMan real-time PCR
TaqMan real-time PCR was performed by using a Perfect Real Time premix (Takara, Japan) containing a high-performance Taq antibody, Takara Ex Taq HS, for hotstart real-time PCR. Primer 3 software was used to design primers and the TaqMan probe with the WSSV whole sequence (GenBank accession no. AF332093). Primers WSSV-RT1 (5′-TTGG TTTCATGCCCGAGATT-3′) and WSSV-RT2 (5′-CCTTGGTCAGCCCCTTGA-3′) produced a fragment of 154 bp after amplification. The TaqMan probe was synthesized and labeled with the fluorescent dyes 5-carboxyfluorescein (FAM) (5′-FAM-TGCTGCCGTCTCCAA-TAMRA-3′). The reaction mixture consisted of DNA aliquot, 200 nM of each primer, 100 nM of each TaqMan probe and 1 × PCR reaction buffer containing DNA polymerase in a final reaction volume of 20 μl. PCR amplification was performed for 4 min at 50°C, followed by 45 cycles of 45 s at 95°C, 45 s at 52°C and 45 s at 72°C. Thermal cycling was performed on an iCycle IQ5 real-time PCR detection system (Bio-RAD, USA).
Statistics
Statistical significance was determined by Student's t-test (two-tailed distribution with a two sample equal variance). P-values of less than 0.05 were considered significant and less than 0.01 were considered very significant.
References
Ravichandran, K. S. & Lorenz, U. Engulfment of apoptotic cells: signals for a good meal. Nature Reviews Immunology 7, 964–974 (2007).
Wu, W. L., Zong, R. R., Xu, J. Y. & Zhang, X. B. Antiviral phagocytosis is regulated by a novel Rab-dependent complex in shrimp Penaeus japonicus. Journal of Proteomic Research 7, 424–431 (2008).
Liu, W. F., Han, F. & Zhang, X. B. Ran GTPase regulates hemocytic phagocytosis of shrimp by interaction with myosin. Journal of Proteomic Research 8, 1198–1206 (2009).
Stuart, L. M. & Ezekowitz, R. L. Phagocytosis and comparative innate immunity: learing on the fly. Nature Reviews Immunology 8, 131–141 (2008).
Stuart, L. M. & Ezekowitz, R. L. Phagocytosis: Elegant Complexity. Immunity 22, 539–550 (2005).
Hoffmann, J. A. & Reichhart, J. M. Drosophila innate imunity: an evolutionary perspective. Nature Immunology 2, 121–126 (2002).
Hoffmann, J. A. The immune response of Drosophila. Nature 12, 33–38 (2003).
Kinchen, J. M. & Ravichandran, K. S. Phagosome maturation: going through the acid test. Nature Reviews Molecular Cell Biology 9, 781–795 (2008).
Bolland, S. A newly discovered Fc receptor that explains IgG-isotype disparities in effector responses. Immunity 23, 2–4 (2005).
Nimmerjahn, F. & Ravetch, J. V. Fc receptors: old friends and new family members. Immunity 24, 19–28 (2006).
Schmidt, R. E. & Gessner, J. E. Fc receptors and their interaction with complement in autoimmunity. Immunology Letters 100, 56–67 (2005).
van der Poel, C. E., Spaapen, R. M., van de Winkel, J. G. & Leusen, J. H. Functional characteristics of the high affinity IgG receptor, FcγRI. Journal of Immunology 186, 2699–2704 (2011).
Nimmerjahn, F. & Ravetch, J. V. Fc-receptors as regulators of immunity. Advance Immunology 96, 179–204 (2007).
Nimmerjahn, F. & Ravetch, J. V. Fcgamma receptors as regulators of immune responses. Nature Reviews Immunology 8, 34–47 (2008).
Ravetch, J. V. & Bolland, S. IgG Fc receptors. Annunal Reviews Immunology 19, 275–290 (2001).
Tanji, T., Hu, X., Weber, A. N. & Ip, Y. T. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Molecular Cell Biology 27, 4578–4588 (2007).
Royet, J., Gupta, D. & Dziarski, R. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nature Reviews Immunology 11, 837–851 (2011).
Leclerc, V. & Reichhart, J. M. The immune response of Drosophila melanogaster. Immunology Reviews 198, 59–71 (2004).
Lemaitre, B. & Hoffmann, J. The host defense of Drosophila melanogaster. Annunal Reviews Immunology 25, 697–743 (2007).
Schmidt, R. L., Trejo, T. R., Plummer, T. B., Platt, J. L. & Tang, A. H. Infection-induced proteolysis of PGRP-LC controls the IMD activation and melanization cascades in Drosophila. FASEB Journal 22, 918–929 (2008).
Ramet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B. & Ezekowitz, R. A. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416, 644–648 (2002).
Park, J. W. et al. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proceedings of the National Academy of Sciences 104, 6602–6607 (2007).
Fedrow, A. L., Ohlen, T. V., Boyle, D., Ganta, R. R. & Chapes, S. K. Use of Drosophila S2 cells as a model for studying Ehrlichia chaffeensis infections. Applied and Environmental Microbiology 74, 1886–1891 (2008).
Underhill, D. M. & Ozinsky, A. Phagocytosis of microbes: complexity in action. Annunal Reviews Immunology 20, 825–852 (2002).
Bergeret, E., Perrin, J., Williams, M., Grunwald, D. & Engel, E. TM9SF4 is required for Drosophila cellular immunity via cell adhesion and phagocytosis. Journal of Cell Science 121, 3325–3334 (2008).
Hashimoto, Y. et al. Identification of lipoteichoic acid as a ligand for draper in the phagocytosis of Staphylococcus aureus by Drosophila hemocytes. Journal of Immunology 183, 7451–7460 (2009).
Stevenson, S. L. S., Foley, E., O'Farrell, P. H. & Johnson, A. D. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biology 4, e4 (2006).
Garver, L. S., Wu, J. & Wu, L. P. The peptidoglycan recognition protein PGRP-SC1a is essential for Toll signaling and phagocytosis of Staphylococcus aureus in Drosophila. Proceedings of the National Academy of Sciences 103, 660–665 (2006).
Miyauchi, M. et al. Phagocytosis plays a dual role in activating dendritic cells; digestive production of active Toll-like receptor ligands and cooperation with Toll-like receptor signaling. Drug Discoveries & Therapeutics 4, 135–143 (2010).
Kubica, M. et al. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS ONE 3, e1409 (2008).
Yu, X., Lu, N. & Zhou, Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biology 6, e61 (2008).
Huber, V. C., Lynch, J. M., Bucher, D. J., Le, J. H. & Metzger, D. W. Fc receptor-mediated phagocytosis makes a significant contribution to clearance of influenza virus infections. Journal of Immunology 166, 7381–7388 (2001).
Bejsovec, A. Flying at the head of the pack: Wnt biology in Drosophila. Oncogene 25, 7442–7449 (2005).
Huelsken, J. & Behren, J. The Wnt signaling pathway. Journal of Cell Science 115, 3977–3978 (2002).
Logan, C. Y. & Nusse, N. The WNT signaling pathway in development and disease. Annual Review of Cell and Developmental Biology 20, 781–810 (2004).
Dobrowolski, R. & De-Robertis, E. M. Endocytic control of growth factor signalling: multivesicular bodies as signalling organelles. Nature Reviews Molecular Cell Biology 13, 53–60 (2012).
Gagliardi, M., Piddini, E. & Vincent, J. P. Endocytosis: A positive or a negative influence on Wnt signalling? Traffic 9, 1–9 (2008).
Gordon, L. R., Gribble, K. D., Syrett, C. M. & Granato, M. Initiation of synapse formation by Wnt-induced MuSK endocytosis. Development 139, 1023–1033 (2012).
Zhu, F., Du, H. H., Miao, Z. G., Quan, H. Z. & Xu, Z. R. Protection of Procambarus clarkii against white spot syndrome virus using inactivated WSSV. Fish Shellfish Immunology 26, 685–690 (2009).
Zhu, F. & Zhang, X. B. Protection of shrimp against white spot syndrome virus (WSSV) with β-1,3-D-glucan-encapsulated vp28-siRNA particles. Marine Biotechnology 14, 63–68 (2012).
Huang, J. X., Sun, B. & Zhang, X. B. Shewanella xiamenensis sp. nov. isolated from coastal sea sediment in China. International Journal of Systematic and Evolutionary Microbiology 60, 1585–1589 (2010).
Berger, S. B. et al. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nature Immunology 11, 920–927 (2010).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research 29, e45 (2001).
Acknowledgements
The authors are grateful to Jian Hong and Guifeng Xiao (Zhejiang University) for transmission electron microscopy and confocal microscopy in the manuscript. This work was financially supported by National Natural Science Foundation of China (31001127), the National Basic Research Program of China (2012CB114403) and Hi-Tech Research and Development Program of China (863 program of China) (2012AA092205-2).
Author information
Authors and Affiliations
Contributions
X.Z. and F.Z. conceived the experiments; F.Z. carried out the experiments; X.Z. and F.Z. analyzed the data. All the authors wrote and reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Zhu, F., Zhang, X. The Wnt signaling pathway is involved in the regulation of phagocytosis of virus in Drosophila. Sci Rep 3, 2069 (2013). https://doi.org/10.1038/srep02069
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02069
This article is cited by
-
MicroRNA-100 is involved in shrimp immune response to white spot syndrome virus (WSSV) and Vibrio alginolyticus infection
Scientific Reports (2017)
-
Nonfunctional ingestion of plant miRNAs in silkworm revealed by digital droplet PCR and transcriptome analysis
Scientific Reports (2015)
-
Role of JAK-STAT signaling in maturation of phagosomes containing Staphylococcus aureus
Scientific Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.