Abstract
Streptococcus pneumoniae pili contribute to adherence and virulence. The regulation of pilus-1 expression is bistable, thus piliated strains contain a variable proportion of pilus-1-non-expressing bacteria. We investigated whether such proportion changes during colonization. Pilus-1-expressing bacteria were quantified in nasopharyngeal washes and pharyngeal tissues from mice that received intranasally bacterial populations with high (H), medium (M) or low (L) pilus-1 expression rates. In nasopharyngeal washes, at early colonization stages, pilus-1 expression rates decreased in H population, while increased in L and M; at later stages, expression rates decreased or remained low. Similar trends were observed in pharyngeal tissues, where, however, at late stages the expression rates were medium-high. In conclusion, pilus-1 is preferentially expressed at early colonization stages, consistently with its role in adhesion, while at later stages the expression is partially switched off. Pilus-1 expression rates observed in clinical isolates in vitro may not reflect the actual rates during colonization/infection.
Similar content being viewed by others
Introduction
Streptococcus pneumoniae colonizes the human nasopharynx and constitutes the etiologic agent of non-invasive and invasive diseases such as otitis media, pneumonia and meningitis, contributing to morbidity and mortality worldwide. Several pneumococcal virulence factors are responsible for its pathogenesis and ability to infect the human host. One of these factors is the pilus, identified in S. pneumoniae in 20061.
Pilus islet-1 (PI-1) contains genes encoding a positive transcriptional regulator (rlrA), the pilus-1 structural subunits (rrgA, rrgB and rrgC) and three pilin-specific sortase1,2. PI-1 is present in about one-third of the clinical isolates3, with variability observed in different epidemiological studies4,5; its prevalence is higher among antibiotic non-susceptible strains3,4. Pneumococcal pilus-1 contributes to both adherence and virulence1,6,7. Interestingly, a recent study reported a significant increase of PI-1 prevalence among clinical isolates, suggesting that expression of the pilus may confer an intrinsic advantage to bacterial transmission8.
Pilus-1 antigens have been proposed as vaccine candidates, exerting high protective efficacy in mice9,10. Pilus-1 expression is bistable11,12, implying that PI-1-positive strains contain variable proportions of pilus-1-expressing and pilus-1-non-expressing bacteria. In vitro stable populations with high (H) or low (L) proportions of pilus-1-expressing bacteria can be obtained by colony selection11. Surprisingly, a chimeric vaccine candidate based on the pilus-1 RrgB subunit (RrgB321) was protective even against L populations, despite the low pilus-1 expression rates at the time of infection, leading to hypothesize for these populations up-regulation of pilus-1 expression in vivo13.
In the present work we analysed the S. pneumoniae pilus-1 expression rates during experimental colonization in mice with selected H or L, or unselected (medium, M) populations of two PI-1-positive strains. “Pilus expression rate” throughout the present paper means the percentage of pilus-1-expressing bacteria for a given PI-1-positive strain/population; it does not refer to the level of pilus-1 expression in pilus-1-expressing bacteria.
Results
Quantification of pilus-1 expression rates in bacterial inputs
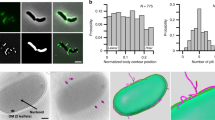
Bacteria administered to mice (input) were analysed by Flow Cytometry (FC) and Indirect Immunofluorescence (IIF). Pilus-1 expression rates for H, M and L bacterial populations grown for the experiments reported in the present paper were as follows: H ≥ 82%; 37% < M < 65%; L ≤ 30%, calculated as the mean of the values quantified by IIF and FC. Figure 1 shows FC and IIF analyses of representative inputs for each bacterial strain/population.
Typical pilus-1 expression rates of each of the S. pneumoniae strains/populations.
The strain analysed is indicated on the top and the population on the right. Quantification of pilus-1 expression rates by FC and IIF are shown on the bottom of the corresponding panel. FC analysis profile: positions of pilus-1-positive or -negative peaks are indicated on the top. IIF: the area of each image is about one third of one of the microscope fields used for quantification; bars = 25 μm. High-brightness images are shown for better readability.
Nasopharyngeal colonization persistence
The bacterial load in nasopharyngeal washes and pharyngeal tissue at the different time-points is summarized in Table 1. It must be pointed out that the pharyngeal mucosa constitutes only part of the mucosa of the mouse upper respiratory tract; thus, the amount of bacteria measured in the pharyngeal tissue homogenate represents a fraction of the total amount of the bacteria adherent to the upper respiratory mucosa. This allows evaluating the differences of pilus expression rates in nasopharyngeal washes and in pharyngeal tissues, but not quantitative comparison between the amounts of bacteria measured in the two specimens, which was out of the aims of this study.
On day 7 post-infection, bacteria were detectable in nasopharyngeal washes of 45% to 100% of mice, depending on the strain/population. Bacteria were also detectable in pharyngeal tissue homogenates, but in fewer mice (from 22% to 62%) than those found S. pneumoniae-positive in nasopharyngeal washes. The mutant 19F_Pc_rlrA on day 7 produced similar colonization than the wild-type strain (Log10 CFU = 3.49 in nasopharyngeal washes, 2.30 in pharyngeal tissue).
Pilus-1 expression is preferentially enhanced at the very early stages of colonization
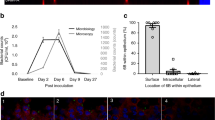
The course of pilus-1 expression rates, summarized in Figure 2, was similar for TIGR4 and 19F. The infection doses (input) and the number of animals are indicated in Table 1.
Pilus-1 expression rates measured in nasopharyngeal washes (a) or pharyngeal tissues (b).
Combination of at least two experiments carried out under the same conditions. Each circle represents the value for a single mouse. Horizontal bars represent the geometric mean for the group with 95% confidence interval. Geometric mean for the first input and geometric means for day 1, 2 and 7 outputs are connected by a dotted line to highlight the changes of pilus-1 expression rates in the course of colonization. The P values, evaluated by two-tailed Mann-Whitney U test, are indicated on the bottom of each graph.
In nasopharyngeal washes (Figure 2 a), on day 1 post-infection, H and L pneumococcal populations changed their pilus-1 expression rates in the direction opposite to that of the input. Indeed, a strong decrease was observed for H populations, while a dramatic increase was seen for L populations (P < 0.001 in both cases). For M populations, pilus-1 expression rates increased, significantly for 19F (P = 0.0008) but not for TIGR4 M (P = 0.1654). On day 2, pilus-1 expression rates tended to decrease (M and L) or remained stable (H populations) with respect to day 1 values.
In the pharyngeal tissue (Figure 2 b), on the first two days post-infection, trends similar to those found in the nasopharyngeal washes were observed, i.e. an increase of pilus-1 expression rates, moderate for M and strong for L populations and a decrease for H populations. The variations were significant for all populations on day 1 and/or on day 2, with the exception of TIGR4 M that did not reach significant differences from the input.
At late stages of colonization, pilus-1 expression is preferentially decreased in nasopharyngeal washes while remaining high in pharyngeal tissue.
On day 7 post-infection, in nasopharyngeal washes (Figure 2 a), pilus-1 expression rates reached low-medium values, with a similar trend for the two strains. In comparison to day 1, pilus-1 expression rates were significantly lower for the M and L populations (P < 0.01 in all cases), while marginally reduced for H populations that had already undergone a significant decrease on day 1. Pilus-1 expression rates were similar among all population/strains with the exception of TIGR4 H that showed slightly but significantly lower values (P = 0.0331).
In the pharyngeal tissue (Figure 2 b), at the last time-point, pilus-1 expression rates were medium-high, with non-significant differences among the populations/strains (P = 0.2752). Interestingly, within each population, pilus-1 expression rates in the pharyngeal tissue on day 7 were higher than those observed in the nasopharyngeal washes (P < 0.0706 for TIGR4 M, < 0.01 for all other strains/populations).
The experiment of colonization with the 19F_Pc_rlrA mutant, constitutively expressing pilus-1 (i.e., input pilus expression rate = 100%), confirmed a substantial difference between the non-adherent (nasopharyngeal washes) and adherent bacteria (pharyngeal tissue homogenate). On day seven, the pilus-1 expression rate in 19F_Pc_rlrA from nasopharyngeal washes was 91%, still close to that of the input. In contrast, 53% of the bacteria harvested from pharyngeal tissue were pilus-1-negative. The CFU count on selective and non-selective plates revealed that 5% of the bacteria from nasopharyngeal washes and 48% of those from pharyngeal tissue homogenates had lost the pMU1328-Pc_rlrA plasmid. These values were consistent with the observed reduction of pilus expression rates.
Specific mucosal antibodies to pilus-1 are undetectable on day 7
No IgG, IgM, or IgA against the pilus-1 RrgB subunit in the naso-pharyngeal washes taken on day 7 were detected by ELISA.
Discussion
The understanding of whether the in vivo expression of pneumococcal pilus-1 undergoes variations may provide valuable information on S. pneumoniae physiology and pathogenesis. In the present study we aimed at detecting variations of pilus-1 expression rates during nasopharyngeal colonization in mice.
An increase of pilus expression rates in both M and L populations was observed at early stage of colonization. This could be due to an advantage conferred to the bacterium by the expression of pilus-1 at this stage of colonization, facilitating the adhesion to host cells1,6. On the other hand, pilus-1 and in particular its backbone subunit RrgB, is highly immunogenic9 and represents a good target for the immune system effectors: thus, its quick switch-off by all populations with the progression of colonization may constitute an effective strategy to reduce the induction and the efficacy of the immune response against pilus-1. Indeed, on the first two days of colonization it is premature to expect specific antibody response in an unprimed host; conversely, at late stages, the antibody-mediated attack could become relevant.
The lack of detectability on day 7 of mucosal Ig against RrgB pilus-1 subunit indicates that the observed reduction of pilus-1 expression rates was not due to selective antibody-mediated killing of pilus-1-expressing bacteria, but rather to a regulation of pilus-1 expression that is preferentially switched off before the host antibody response is mounted. The mechanisms of such regulation are presently unknown and require further investigation; however, they could include a response to environmental factors. Indeed, the Mn2+ concentration equivalent to that present in the oral mucosa environment has been reported to repress the transcription of some pneumococcal genes including rlrA14, which positively regulates pilus-1 expression11.
At late stages of colonization, pilus-1 expression rates were higher in the pharyngeal tissues (containing the more adherent bacteria) than in the nasopharyngeal washes (containing the less adherent bacteria), confirming the importance of S. pneumoniae pili in adherence1,6. The results of colonization with the 19F_Pc_rlrA mutant, constitutively expressing pilus-1, give further explanation of the mechanisms that regulate the changes on pilus-1 expression. While the loss of the plasmid observed in 5% of bacteria from nasopharyngeal washes can be considered very marginal, the loss in 48% of bacteria from pharyngeal tissue indicates selective pressure toward non-piliated phenotype at the tissue level. On the other hand, the relatively high pilus-1-expression rates observed in the tissue for the wild-type strains indicate an opposite trend, toward pilus-1 expression. The recent observation that the RrgA pilus-1 subunit enhances the non-opsonic uptake of bacteria by macrophages, at the same time promoting the survival within the phagocytes15, can help to explain what we observed at the tissue level. Here, pilus-1-expressing bacteria could be quickly sequestered by phagocytes, in the frame of the innate immunity defense; on the other hand, bacteria would tend to switch on pilus-1 expression, since it is advantageous to the colonization/infection strategy. This would lead to reach the equilibrium that we observed between pilus-1-expressing and pilus-1-non-expressing bacteria at the tissue level. It has been recently shown that pilus-1, through its RrgA subunit, favours the S. pneumoniae spreading from the mouse respiratory tract or peritoneum to the bloodstream15: the presence of a reservoir of piliated bacteria at the upper respiratory tissue level and the bacterial ability of modulating pilus expression during colonization that we describe in the present study could play a relevant role in this pilus-dependent S. pneumoniae invasion strategy.
We have previously reported the high protective efficacy of a pilus-1-based vaccine candidate in mice9,10. Remarkably, we have recently observed the efficacy of a pilus-1-based vaccine in murine sepsis and pneumonia models even against L populations13. We hypothesized that in the host the L population switched-on pilus-1 expression, making the bacteria vulnerable to the immune response against pilus-1 induced by vaccination. This hypothesis is now sustained by the present observation that pilus-1 expression rates of S. pneumoniae L populations transiently increase in vivo, further supporting the use of the pilus-1-based vaccine against all piliated S. pneumoniae strains.
In conclusion, our data demonstrate that S. pneumoniae pilus-1 expression can be switched on and off in vivo and provide evidence that high pilus-1 expression is preferentially switched on at early stages of colonization (suggesting that pilus-1 may confer an advantage at this stage), while low-medium rates are preferred at late stages. The latter phenomenon may represent a micro-adaptation mechanism by which piliated S. pneumoniae strains tend to avoid the host immune response, while maintaining a reservoir of piliated bacteria. The two strains tested, TIGR4 (invasive) and 19F (non-invasive), showed similar behaviour, suggesting that the variations of pilus-1 expression rates are not related to the invasiveness of the strain, at least in this mouse colonization model. Further studies to evaluate the changes of pilus-1 expression rates in pneumonia and in systemic infection models could reveal possible relationships between pilus-1 expression and different states of infection and disease.
For several Gram-positive bacteria, heterogeneous expression of pili has been reported and variations of pilus expression rates have been observed in vitro in response to environmental factors16. To our knowledge the present study is the first to provide evidence of such variations in vivo, making evident that the pilus-1 expression rate observed in a cultured clinical isolate may not reflect the actual rate in the course of colonization/infection.
Methods
Bacterial strains and culture
H, L, or M populations of TIGR4 (serotype 4) and 19F-Taiwan 14 (serotype 19F, from here referred to as 19F) PI-1-positive S. pneumoniae strains were used. Additionally, the 19F mutant 19F_Pc_rlrA, which constitutively expresses the regulator RlrA and therefore has 100% pilus-1 expression rate, was used. Bacteria were grown 24 h at 37°C under 5% CO2 atmosphere on Tryptic Soy Agar (TSA, Difco) plates containing colistine 10 mg/l, oxolinic acid 5 mg/l and 5% defibrinated sheep blood. Bacteria from plates were used to inoculate liquid cultures in Tryptic Soy Broth (TSB, Difco). Growth was carried out statically at 37°C under 5% CO2 atmosphere until A600 = 0.25 was reached. Bacteria were then harvested by centrifugation at 3,500 r.c.f. for 20 min at 4°C, resuspended in saline and used within 1 h for intranasal administration. Bacteria for immunization (M populations) were heat-inactivated for 1 h at 60°C and stored at 4°C until use. The colony forming units (CFU) per ml were determined by seeding serial dilutions on plates and counting the colonies after overnight culture carried out as above.
Construction of 19FPcrlrA mutant
The pMU1328 recombinant plasmid (pMU1328-Pc_rlrA)17 was obtained as previously described18. Briefly, the rlrA gene was amplified on the TIGR4 genomic DNA (rlrA Forward GTGCGTGGATCCATGCTAAACAAATACATTGAAAAA, rlrA Reverse CAGCGTGTCGACCTTTTTGTGTGTAGACAGTACGAT), while the erythromycin resistance (ermB) constitutive promoter (Pc) was amplified on the pMU1328 plasmid (Pc Forward GTGCGTGAATTCGAAACAGCAAAGAATGGCGGAAAC, Pc Reverse CAGCGTGGATCCGTAATCACTCCTTCTTAATTACAA). The obtained PCR products were digested with Bam HI – SaI I for rlrA and EcoRI – Bam HI for Pc and cloned into the complementation plasmid pMU1328 digested with EcoRI – BamHI. The 19F strain was then transformed with conventional methods19 by using the obtained plasmid containing an erythromycin resistance marker17. Transformants were selected on plates supplemented with 100 μg/ml erythromycin and resulted stable upon repeated culture cycles. The plasmid presence was confirmed by PCR and the expression of pilus-1 proteins was confirmed by flow cytometry (FC) (not shown). The evaluation of possible plasmid loss was made by quantitative comparison of growth on plates with 100 μg/ml erythromycin (selective) and on plates with 1 μg/ml erythromycin (non-selective).
Animal treatments and sample preparation
Animal experiments were done in compliance with the current European guidelines and approved by the internal Animal Ethics Committee (AEC 200911 and AEC-2011-06).
Colonization in mice was achieved according to Wu and colleagues20, with slight modifications. Non-anesthetized female, 8-week-old, BALB/c mice, received into each nostril 5 μl of bacterial suspension. The infection doses and the number of animals are indicated in Table 1. At least two experiments for each bacterial population (H, L or M) were repeated under similar conditions. Groups of animals were euthanized 1, 2 and 7 days post-infection; the animals colonized with the mutant 19FPcrlrA were sampled only on day 7. Nasopharyngeal washes were obtained by resecting the trachea and flushing 0.5 ml saline from the trachea through the nostrils. After the washes, to analyse the bacteria remaining adherent to the mucosa, pharyngeal tissues were excised and homogenized in 5 ml saline using gentleMACS dissociator (Miltenyi Biotec). The homogenate was centrifuged at 3,500 r.c.f. for 20 min at 4°C and resuspended in 0.5 ml saline. For CFU count, aliquots of both nasopharyngeal washes and concentrated pharyngeal tissue homogenate were cultured in plates as above. For immunostaining, specimens were inactivated in 70% ethanol for 60 min at R.T., pelleted at 6,000 r.c.f. for 10 min at 4°C, resuspended in 20 μl saline and kept at 4°C until use.
Single animals were analysed with the exception of the experiment where colonization was established with the mutant 19F_Pc_rlrA, in which pooled specimens from 4 mice were analysed.
Antibody preparation
Anti-TIGR4 and -19F sera were raised in New Zealand rabbits using heat-inactivated bacteria. Rabbits received subcutaneous injection of the equivalent of 109 CFU on day 0, 21 and 35, along with Freund's adjuvant and were bled on day 49. Anti-RrgB sera were raised both in guinea pigs and in rabbits administering three doses of 100 μg of recombinant RrgB with the above-described procedure. Recombinant RrgB was expressed in E. coli and purified as previously described9.
Analysis of pilus-1 expression rates
The first time-point analysed was set at one day post-infection, to avoid that, at shorter time-points, the analysis of pilus-1 expression rates reflected the in vitro phenotype rather than possible modifications due to colonization. Indeed, when setting up the study, administration of bacteria labelled with carboxyfluorescein diacetate N-succinimidyl ester (CFDA-SE) revealed that 3 hours post-infection most bacteria were still fluorescent (not shown), indicating the presence of dead or non-replicating cells from the input. Conversely, one day post-infection, bacteria were non-fluorescent, indicating that a sufficient number of replications occurred (not shown).
For cultured bacteria, pilus-1 expression rates were quantified both by flow cytometry (FC) and indirect immunofluorescence (IIF). Bacteria directly harvested from nasopharyngeal washes and pharyngeal tissues did not reach sufficient amount to allow either FC analysis or mRNA purification in order to quantify the transcription of pilus-1 genes (not shown). Thus, on freshly harvested biological specimens, pilus-1 expression rates were quantified by IIF.
Indirect immunofluorescence (IIF)
Pilus-1 expression rates were quantified in specimens positive for CFU count. Five μl of each sample were spotted onto poly-L-lysine-coated microscope slides and let dry at 37°C. Slides were rehydrated and saturated with 1% BSA, then incubated with either anti-TIGR4 or anti-19F (1 : 2,000) together with guinea pig anti-RrgB (1 : 500) for 60 min at R.T., followed by Alexa-Fluor-488-conjugated goat anti-rabbit and Alexa-Fluor-568-conjugated goat anti-guinea pig (1 : 200, Invitrogen). Slides were observed with epi-fluorescence microscope with 40x lens. Total and pilus-1-positive bacteria were blind counted in ≥ 4 microscope fields to reach ≥ 100 total bacteria.
Flow cytometry (FC)
Aliquots of bacterial suspensions were incubated with rabbit anti-RrgB (1 : 400) followed by incubation with secondary FITC-conjugated anti-rabbit antibody (1 : 100, Jackson Laboratories) and fixation with 2% formaldehyde. Pilus-1 expression rates were analysed by using a FACSCalibur cytometer (Becton Dickinson). The position of pilus-1-negative peak was determined by analyzing bacteria treated with the secondary antibody only. For each sample 10,000 events were recorded and the percentage of pilus-1-positive bacteria within each sample was estimated with the CellQuest software (Becton Dickinson).
Antibody titration by Enzime-Linked Immunosorbent Assay (ELISA)
Pooled nasopharyngeal washes, harvested on day 7, were 0.2 μm filtered on filters saturated with 0.1% BSA. Nasopharyngeal washes from untreated mice were used as a control.
The presence of specific mucosal Ig against RrgB, which is the most immunogenic pilus-1 subunit9, was analysed. 96-well plates (Nunc) were coated with 0.2 μg/well of recombinant RrgB, then saturated with 1% BSA in PBS and subsequently incubated for 2 h at 37°C with 100 μl of 1 : 2 dilution of washes. Bound IgG, IgA or IgM were detected with specific alkaline phosphatase-conjugated antibodies (1 : 2,000, Southern Biotech) incubated for 1 h at 37°C. The colour was developed by adding p-nitro-phenyl-phosphate substrate and read at 405 nm in an ELISA reader.
Statistical analysis
Differences of pilus-1 expression rates between input and output at each time-point and those between day 1 and day 7 of each population, were evaluated by two-tailed Mann-Whitney U test. Multiple comparisons among the various populations on day 7 were done by Kruskal-Wallis one-way ANOVA. Significance limit was set at P ≤ 0.05.
References
Barocchi, M. A. et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA 103, 2857–2862 (2006).
LeMieux, J., Hava, D. L., Basset, A. & Camilli, A. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniaerlrA pathogenicity islet. Infect. Immun. 74, 2453–2456 (2006).
Moschioni, M. et al. Streptococcus pneumoniae contains 3 rlrA pilus variants that are clonally related. J. Infect. Dis. 197, 888–896 (2008).
Sjostrom, K. et al. Clonal success of piliated penicillin nonsusceptible pneumococci. Proc. Natl. Acad. Sci. USA 104, 12907–12912 (2007).
Selva, L. et al. Prevalence and clonal distribution of pcpA, psrP and pilus-1 among pediatric isolates of Streptococcus pneumoniae. PLoS One 7, e41587 (2012).
Nelson, A. L. et al. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 66, 329–340 (2007).
Hilleringmann, M. et al. Molecular architecture of Streptococcus pneumoniae TIGR4 pili. EMBO J. 28, 3921–3930 (2009).
Regev-Yochay, G. et al. Re-emergence of the type 1 pilus among Streptococcus pneumoniae isolates in Massachusetts, USA. Vaccine 28, 4842–4846 (2010).
Gianfaldoni, C. et al. Streptococcus pneumoniae pilus subunits protect mice against lethal challenge. Infect. Immun. 75, 1059–1062 (2007).
Harfouche, C. et al. RrgB321, a fusion protein of the three variants of the pneumococcal pilus backbone RrgB, is protective in vivo and elicits opsonic antibodies. Infect. Immun. 80, 451–460 (2012).
De Angelis, G. et al. The Streptococcus pneumoniae pilus-1 displays a biphasic expression pattern. PLoS One 6, e21269 (2011).
Basset, A. et al. Expression of the type 1 pneumococcal pilus is bistable and negatively regulated by the structural component RrgA. Infect. Immun. 79, 2974–2983 (2011).
Moschioni, M. et al. Immunization with the RrgB321 fusion protein protects mice against both high and low pilus-expressing Streptococcus pneumoniae populations. Vaccine 30, 1349–1356 (2012).
Johnston, J. W., Briles, D. E., Myers, L. E. & Hollingshead, S. K. Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect. Immun. 74, 1171–1180 (2006).
Orrskog, S. et al. Pilus adhesin RrgA interacts with complement receptor 3, thereby affecting macrophage function and systemic pneumococcal disease. MBio. 4, e00535–12 (2012).
Danne, C. & Dramsi, S. Pili of Gram-positive bacteria: roles in host colonization. Res. Microbiol. 163, 645–658 (2012).
Achen, M. G., Davidson, B. E. & Hillier, A. J. Construction of plasmid vectors for the detection of streptococcal promoters. Gene 45, 45–9 (1986).
Lo Sapio, M., Hilleringmann, M., Barocchi, M. A. & Moschioni, M. A novel strategy to over-express and purify homologous proteins from Streptococcus pneumoniae. J. Biotechnol. 157, 279–286 (2012).
Alloing, G., Martin, B., Granadel, C. & Claverys, J. P. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29, 75–83 (1998).
Wu, H. Y. et al. Establishment of a Streptococcus pneumoniae nasopharyngeal colonization model in adult mice. Microb. Pathog. 23, 127–137 (1997).
Acknowledgements
Animal treatments were carried out by Marco Tortoli, Elena Amantini and Luigi Manganelli. Raffaella Cecchi performed the excision of pharyngeal tissues. We thank Vega Masignani for the support provided as a project leader of the S. pneumoniae vaccine project and for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
All authors (L.P., G.D.A., E.B., M.A.B., G.D.G., M.M. and P.R.) contributed to interpret the results and participated in writing the manuscript. In addition: L.P. took care of in vivo experiments and IIF analysis and quantification; G.D.A. performed the selection of bacterial populations and FC analyses; E.B. contributed to the in vivo experiments and microbiological analyses; M.M. and P.R. conceived the study; P.R. took care of the experimental design and of results elaboration and wrote the paper.
Ethics declarations
Competing interests
All authors are Novartis employees, with the exception of E.B., who was Novartis temporary employee when the study was carried out.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Pancotto, L., De Angelis, G., Bizzarri, E. et al. Expression of the Streptococcus pneumoniae pilus-1 undergoes on and off switching during colonization in mice. Sci Rep 3, 2040 (2013). https://doi.org/10.1038/srep02040
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02040
This article is cited by
-
Comparison of specific in-vitro virulence gene expression and innate host response in locally invasive vs colonizer strains of Streptococcus pneumoniae
Medical Microbiology and Immunology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.