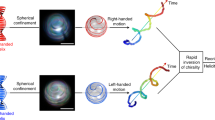
Abstract
We report the discovery of an entirely new three-dimensional (3D) swimming pattern observed in human and horse sperms. This motion is in the form of ‘chiral ribbons’, where the planar swing of the sperm head occurs on an osculating plane creating in some cases a helical ribbon and in some others a twisted ribbon. The latter, i.e., the twisted ribbon trajectory, also defines a minimal surface, exhibiting zero mean curvature for all the points on its surface. These chiral ribbon swimming patterns cannot be represented or understood by already known patterns of sperms or other micro-swimmers. The discovery of these unique patterns is enabled by holographic on-chip imaging of >33,700 sperm trajectories at >90–140 frames/sec, which revealed that only ~1.7% of human sperms exhibit chiral ribbons, whereas it increases to ~27.3% for horse sperms. These results might shed more light onto the statistics and biophysics of various micro-swimmers' 3D motion.
Similar content being viewed by others
Introduction
Locomotion in an aqueous environment plays an important function in the lives of many micro-organisms, such as bacteria, protists and sperms. Without this kind of self-propelled swimming act, these micro-organisms cannot actively find food, escape predators, or produce offsprings to sustain their species. Understanding the swimming patterns of these micro-organisms and the underlying biophysical processes is important to advance our existing knowledge in microbiology. To resolve the spatial and temporal details of these tiny (e.g., 1–50 μm) micro-organisms' fast locomotion (with speeds of for example 50–500 μm/sec), lens-based optical microscopes with high-speed cameras (e.g., >60 frames per sec, FPS) have been typically employed to digitally record their trajectories. Due to the restricted imaging volume of microscope objective lenses (having a limited field-of-view and depth-of-field), most of the earlier studies on these micro-organisms' locomotion have been limited to their two-dimensional (2D) dynamics1,2,3,4,5,6,7,8,9,10,11. Relatively recently, three-dimensional (3D) optical tracking techniques have also emerged, enabling the observation of various unique 3D swimming patterns of these micro-organisms, including for example helical, star-spin and planar motion12,13,14,15,16,17,18,19,20,21,22,23. Among these recent techniques, computational lensfree on-chip microscopy tools are especially well suited to analyze rare 3D swimming events that occur at the micro-scale because, unlike their lens-based counterparts, they have a much higher throughput to track a large number of individual trajectories in 3D14,16,23,24.
Using a dual-angle and dual-color illumination based computational on-chip imaging platform23, here we report the discovery of an entirely new swimming pattern observed in human and horse sperms. This rare 3D motion is in the form of ‘chiral ribbons’ that resemble the chiral structures observed in several other biological systems25,26,27,28 (see Fig. 1 for a 3D illustration of chiral ribbons). In this swimming pattern, left-to-right and right-to-left planar swing of the sperm head occurs on an osculating plane creating in some cases a ‘helical ribbon’ and in some others a ‘twisted ribbon’. Quite interestingly, the twisted ribbon trajectory mathematically defines a minimal surface, exhibiting zero mean curvature for all the points on its surface, which will be proved and further discussed in our Discussion Section. These dynamic trajectories defined by chiral ribbons cannot be represented or understood by already known patterns of sperms of various species or other micro-swimmers1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,23. The discovery of these unique 3D patterns is enabled by lensfree holographic on-chip imaging and tracking of ~1,000–1,600 sperms (per experiment) at a frame rate of >90–140 frames per second over a large sample volume of e.g., ~9 μL, which is more than an order-of-magnitude larger compared to a lens-based optical microscope. Our high-throughput imaging experiments revealed that only ~1.7% of human sperms (out of 24,090 individual sperm trajectories) exhibit chiral ribbons, whereas the same percentage increases to ~27.3% for horse sperms (out of 9,625 individual sperm trajectories). We also observed that the majority (~65%) of the chiral ribbons displayed by human sperms are right-handed, whereas ~85% of the chiral ribbons observed in horse sperms exhibit left-handed rotations. We believe that these results could help us better understand the statistical behavior of various micro-swimmers and might shed more light onto the biophysics of their 3D motion.
Results
Using a dual-view and dual-wavelength holographic on-chip microscopy platform (see Fig. 2), we initially imaged and reconstructed 3D spatio-temporal trajectories of horse sperms within large sample volumes (~9 μL) at ~140 FPS (refer to the Methods Section for details). Based on these experiments, we observed that horse sperms display a unique ‘chiral ribbon’ pattern (see Fig. 3–4), which occurs with a frequency of ~27.3% among 9,625 individual 3D trajectories that we reconstructed using our high-throughput on-chip imaging platform. These horse sperms that follow a chiral ribbon swimming pattern have locally planar lateral displacements as if they are confined in 2D. In contrast to regular planar trajectories, however, in a chiral ribbon the plane on which the lateral displacements occur rotates continuously around a central axis as the sperm is moving forward, forming a ‘twisted’ ribbon within e.g., a ~0.7-sec long segment of the sperm trajectory (see Fig. 3(b,e)). Not only is this twisted ribbon trajectory quite tight with a typical side-to-side displacement of ~5 μm, but also it beats rather fast, crossing over the central axis roughly 30 times within a second, making it rather challenging to observe with other 3D optical tracking techniques12,13,14,15,16,17,18,19,20,21,22,29,30 due to the tight requirements in 3D localization accuracy and video frame rate.
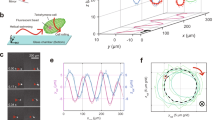
Dual-view and dual-wavelength lensfree 3D tracking of sperms on a chip.
(a) The picture of the imaging system. Two partially-coherent light sources (LED-coupled multimode fibers, core size: 400 μm) illuminate the observation chamber from two different angles with two different wavelengths (vertical one: 625 nm; oblique one at 45°: 470 nm; bandwidth ~20 nm). A CMOS image sensor records the dual-view lensfree holograms that encode the position information of each sperm. (b) The close-up image of an observation chamber that is placed on the top of the CMOS image sensor. (c) The reconstructed 3D spatio-temporal trajectories of 998 horse sperms that were tracked inside a volume of ~9.0 μL at a frame rate of 143 FPS. The inset in (c) shows the trajectories reconstructed inside a sub-volume of the lensfree imaging system. The time position of each track point is encoded by its color (see the color bar).
The chiral ribbon patterns displayed by horse sperm trajectories.
(a) and (d) are two chiral ribbon trajectories recorded over a duration of 4.6 sec. (b) and (e) are digitally zoomed segments (~0.7-sec long each) of the two ribbon trajectories shown in (a) and (d), respectively. Both of the trajectories in (a) and (d) have left-handed twisting and form left-handed helical ribbons. (c) and (f) are two simulated trajectories that were generated by equation 2 to match the measured chiral ribbon trajectories shown in (a) and (d), respectively (see Table 1 for the simulation parameters). The magenta ribbon surfaces in (b) and (e) designate the osculating planes of the twisting ribbon trajectories. The inset in each panel represents the lateral displacement of the straightened sperm trajectory (front view). The arrows indicate the directions of the sperms' forward movement. The time position of each track point is encoded by its color (see the color bar). See Supplementary Movies S1 and S2 for the spatio-temporal evolution of the ribbon trajectories shown in (a) and (d), respectively.
A 5.0-sec long 3D trajectory showing the transitions between different swimming patterns of a horse sperm.
This trajectory switched from a right-handed chiral ribbon pattern to a simple planar swimming pattern when the sperm encountered the bottom glass surface of the observation chamber. (b), (c) and (d) illustrate digitally zoomed segments (~0.7-sec long each) of the whole sperm trajectory shown in (a). The inset in each panel represents the lateral displacement of the straightened sperm track segment (front view). The arrows indicate the directions of the sperm's forward movement. The time position of each track point is encoded by its color (see the color bar). See Supplementary Movie S3 for the spatio-temporal evolution of this trajectory.
Further investigation of these chiral ribbon swimming patterns in horse sperm samples revealed that in longer durations of observation (>2.1 sec), we can also observe spatio-temporal super-structures that mimic a different type of a chiral ribbon, namely a “helical ribbon” (see e.g., Fig. 3(a,d) and Supplementary Movies S1–S2). With this type of super-structure, the osculating plane of the sperm trajectory (i.e., the beating of the sperm head) not only twists as the sperm moves forward, but also forms a 3D helix in the form of a chiral ribbon (see for example the left-handed twisting planes that form the left-handed helical ribbons shown in Fig. 3(a) and (d); or Supplementary Movies S1 and S2, respectively). We should emphasize that such a helical ribbon trajectory should not be confused with already-known helical trajectories of sperms23, where the sperm head defines a simple helix, not a 3D ribbon (i.e., without any chiral planar motion). As a matter of fact, these chiral ribbon swimming patterns cannot be represented by already known patterns of sperms or other micro-swimmers and are reported in the literature for the first time, enabled by our high-throughput on-chip imaging platform. The same lensfree imaging system also permits us to track each one of these ~1,000 horse sperms (per experiment) within a large sample volume of ~9 μL across a time window of e.g., ~7–10 sec (at ~140 FPS) and analyze the spatio-temporal transitions of their swimming patterns into or out of the chiral ribbon pattern. Figure 4 and Supplementary Movie S3 illustrate such a swimming pattern transition observed when an individual horse sperm encountered the bottom glass surface of our observation chamber, where its 3D trajectory switched from a right-handed chiral ribbon pattern to a simple planar swimming pattern. This example illustrates that boundary confinement could be an influential factor in the transitions of sperm swimming patterns.
We also statistically quantified the 3D motion dynamics, such as Rate of Twisting (RTW), Linearity (LIN), Straight-Line Velocity (VSL), Curvilinear Velocity (VCL) and Amplitude of Lateral Head Displacement (ALH) of the chiral ribbon trajectories observed in horse sperms (see Fig. 5 and the Methods Section for details). For this statistical analysis, we digitally processed 2,625 individual chiral ribbon patterns (out of a total of 9,625 trajectories, each ~0.7 sec long) to reveal that ~85% of the ribbon horse sperm trajectories prefer left-handed twisting over right-handed twisting (see Fig. 5(c)), exhibiting a side-to-side displacement of 1–12 μm and a twisting rate (RTW) of 0.5–22 rad/sec.
Dynamic swimming parameters of 2,625 chiral ribbon trajectories (out of a total of 9,625 motile horse sperms, each ~0.7 sec long).
Color bar represents the relative density of data points in each graph. Magenta lines enclose 90% of the chiral ribbon trajectories presented in each panel. VSL: straight-line velocity. VCL: curvilinear velocity. ALH: amplitude of lateral head displacement. LIN: linearity. RTW: rate of twisting. A chiral ribbon trajectory with RTW > 0 (RTW < 0) is defined as right-handed (left-handed). These measurements were all made with horse sperms suspended in equine semen extender (BotuSemen) after >30 min of incubation.
After discovering chiral ribbon trajectories in horse sperm samples, we also searched for similar 3D swimming patterns in human sperms (see the Methods Section). For this end, we digitally processed 24,090 individual human sperm trajectories to find out that only ~1.7% of the time chiral ribbons were formed (see e.g., Fig. 6 and Supplementary Movie S4 for a twisted ribbon pattern). This lower percentage also partially explains why this twisted ribbon type of swimming pattern remained unidentified in earlier studies and was in fact broadly classified as part of the “typical”23 trajectories for human sperms (see Fig. 7). Despite their low percentages, we still observed that right-handed twisting in human sperm ribbons is more frequent than left-handed twisting (1.1% vs. 0.6% of 24,090 trajectories), which is quite the opposite of what is observed with horse sperm ribbons. Another interesting difference between the chiral ribbons of these two species is that human sperms, even in longer observation time windows, still follow twisted ribbons rather than helical ribbons (see e.g., Fig. 6(a)).
(a) A 4.4-sec long human sperm trajectory that forms a twisted ribbon.The magenta ribbon surface in (b) designates the right-handed osculating plane of the trajectory. The insets in the panels represent the lateral displacement of the straightened sperm trajectories (front view). The arrows indicate the directions of the sperms' forward movement. The time position of each track point is encoded by its color (see the color bar). See Supplementary Movie S4 for the spatio-temporal evolution of the sperm trajectory shown in (a).
The statistics of human sperm chiral ribbon swimming patterns compared to other swimming patterns observed in human sperms.
The right-handed and left-handed chiral ribbon patterns (excluding hyperactivated ribbons) account for 1.1% and 0.6% of all the 24,090 human sperm trajectories that we reconstructed, respectively. These measurements were made in human sperm baseline medium (artificial HTF) after >2 h of incubation23.
Discussion
Mathematically, the equation of a ‘chiral ribbon’ surface s can be broadly defined as:

where 0 ≤ l ≤ L,  , L is the length of the chiral ribbon that is aligned along the z-direction, D and rh are the width and the radius of the chiral ribbon, respectively, Ph is the pitch of the chiral ribbon (defined as positive for a right-handed ribbon and negative for a left-handed ribbon) and θh is the offset angle of the chiral ribbon. This chiral ribbon equation can form a “helical” or a “twisted” ribbon when rh is significantly larger than zero or close to zero, respectively.
, L is the length of the chiral ribbon that is aligned along the z-direction, D and rh are the width and the radius of the chiral ribbon, respectively, Ph is the pitch of the chiral ribbon (defined as positive for a right-handed ribbon and negative for a left-handed ribbon) and θh is the offset angle of the chiral ribbon. This chiral ribbon equation can form a “helical” or a “twisted” ribbon when rh is significantly larger than zero or close to zero, respectively.
The chiral ribbons observed in our sperm tracking experiments (see e.g., Figs. 3(a) and 3(d)) can be mathematically reproduced by adding a periodic lateral oscillation (e.g., sinusoidal) along the ribbon surface that is defined by equation 1, i.e.,

where νz is the forward-moving velocity along the z direction, ωh = 2π(νz/ph) is the angular velocity of the helical ribbon (positive for a right-handed ribbon and negative for a left-handed one), rb is the radius of the sperm beating waveform, ωb and θb are the angular velocity and the offset angle of the same beating waveform. Two examples of simulated sperm trajectories that are computed based on equation 2 are shown in Fig. 3(c) and 3(f), with their parameters (Table 1) tuned to match the measured chiral ribbon trajectories of Fig. 3(a) and 3(d), respectively. Note that the osculating planes of the measured sperm trajectories reported in Fig. 3(a) and 3(d) are in very good agreement with our theoretical trajectories shown in Fig. 3(c) and 3(f), respectively, both calculated based on equation 2 with the parameters of Table 1.
Interestingly, when rh = 0, the “twisted” ribbon surface exactly becomes a minimal surface in the form of a helicoid (which should not be confused with a helix)31. Minimal surfaces minimize their surface area with respect to a particular boundary and also have local mean curvature equal to zero at all points on their surface. Besides a helicoid, a simple plane and a catenoid also define minimal surfaces. The mean curvature (H) of a chiral ribbon surface can be computed from the partial derivatives of s with respect to its parameters, l and a and it can be written as32:

where subscripts indicate partial derivatives (e.g.,  ) and
) and  is the unit normal vector. For the chiral ribbon surfaces defined by equation 1, the mean curvature can be calculated as:
is the unit normal vector. For the chiral ribbon surfaces defined by equation 1, the mean curvature can be calculated as:

As the term inside the square root is strictly positive in equation 4, H = 0 and s is a minimal surface if and only if rh = 0, proving that an ideal twisted ribbon indeed defines a minimal surface.
Note that minimal surfaces are also naturally found in a wide range of other fields. For example, minimal surfaces (in the form of catenoids) describe the shape soap films adopt on a wire frame33. Additionally, nano-catenoid minimal surfaces have recently been shown to describe the shape of self-assembled liquid nanolenses that can spontaneously form around nanoparticles or viruses adhered to a substrate34. As another related example, one of the phases of di-block copolymer mixtures, the gyroid phase, is also a minimal surface35. Here the gyroid surface describes the interphase boundary between the two nanoscopic domains of the polymer. Other materials whose structure can be described as minimal surfaces include inorganic crystal structures, biological cell membranes and in certain cases, the structure of folded proteins32,36.
We should emphasize that the actual cause and the biological function of these chiral ribbon swimming patterns are not clear and should be the subject of future work. In many of the situations where minimal surfaces arise, a common theme is that of energy minimization. Hence, the fact that different modes of sperm locomotion (e.g., twisted ribbon as well as simple planar trajectories) follow a minimal surface may also indicate energy minimization corresponding to maximal propulsion efficiency in these modes. However, significantly more investigation into the hydrodynamics and biomechanics of the sperm motion would be required to validate this proposition. Another related speculative cause is that a small asymmetry in the shape or the location of the center-of-mass of the sperm head might generate a net torque through the surrounding fluid when the sperm tail is pushing its body forward. Similar phenomena have been discussed previously using hydrodynamics numerical simulations to partially explain regular helical trajectories (not chiral ribbons) of sperm cells37. Based on such a structural asymmetry, the local movement of the sperm head can still be governed by the planar beating of its tail, however the whole body can gradually rotate due to a net torque and the sperm trajectory can slowly deviate from the central axis of the beating. Once the whole sperm body finishes a full rotation, the trajectory would also complete a cycle of the helical ribbon. As for the biological function of such a chiral ribbon swimming pattern, it might possibly be similar to what has been proposed for regular helical trajectories38,39,40; in other words, these chiral swimming patterns could potentially help reorienting the motion of the micro-organisms into the direction of stimulus gradient due to for example the concentration of a chemo-attractant or just the environmental temperature distribution. These chiral ribbons that we observed, compared to simple planar trajectories, might permit sperms to sense such gradient profiles over larger 3D volumes, which could then help detection of weaker stimuli, assisting in e.g., sperms' chemotaxis41,42 or thermotaxis41,43 behavior. These and other possible hypotheses would need to be further studied in greater detail to better understand the origins and possible functions of chiral ribbon swimming patterns in general.
Methods
Preparing and incubating sperm suspension
Frozen sperm specimens of a Warmblood stallion were obtained from the Center for Equine Health at the University of California, Davis. Before freezing, fresh semen specimens were first diluted to a concentration of 50 million sperms per ml with equine semen extender (E-Z Mixin BFT, Animal Reproduction Systems) and then were centrifuged at 400 G for 15 min. After centrifugation the pellet was re-suspended in freezing extender (E-Z Freezin Equine Semen Extender, Animal Reproduction Systems) with a final concentration of 400 million sperms per ml. The processed sperms specimens were packaged in 0.5 ml straws and frozen in a programmable freezer. When the straws had reached −150°C they were plunged in liquid nitrogen for storage.
To prepare the horse sperm suspension for imaging, the frozen specimens were first thawed at 38°C water bath for 30 sec and then rehydrated for 15 min by mixing with equine semen extender (BotuSemen, Nidacon, Sweden) by a ratio of 1:1. After rehydration, gradient density centrifugation with isotonic density medium (Equipure, Nidacon, Sweden, 200 g for 30 min) was used to concentrate the motile sperms within the semen specimens. The centrifuged sperm pellet was re-suspended with the same equine semen extender at a concentration of ~1 million sperms per ml (>50% motile) and then incubated for another 30 min. Right before lensfree on-chip imaging experiments, ~25 μL of the sperm suspension was put into a disposable observation chamber prepared by taping a laser-cut Acetal film (~0.15 mm thick) between two pieces of No. 1 cover slips.
The methods and related procedures for preparing and incubating human sperm suspensions have been explained in detail in our previous work23.
Dual-view and dual-wavelength lensfree on-chip holographic imaging and tracking set-up
A dual-view and dual-wavelength lensfree on-chip holographic imaging setup, as illustrated in Fig. 2, was utilized to record the 3D movement of sperms. Two partially-coherent light sources (LED-coupled multimode fibers, core size: 400 μm) illuminated the observation chambers from two different angles with two different wavelengths (vertical one: 625 nm; oblique one at 45°: 470 nm; bandwidth ~20 nm). When recording the 3D movement of sperms, the observation chamber was placed directly on the top of the protection glass of our CMOS (Complementary Metal-Oxide-Semiconductor) image sensor (Aptina MT9P031STC, 5 megapixels, 2.2 μm pixel size, monochrome; see Fig. 2(b)). The power of this image sensor chip was cut off between video acquisition sessions to maintain the temperature of the sperm observation chamber at ~37–39°C.
The frame rate of the computational imaging system used in this manuscript was raised to 143 FPS to oversample the faster beating of horse sperms (beat-cross-frequency, BCF: ~30 Hz), whereas it was operated at ~92 FPS for imaging of human sperms23. Such high frame rates reduced the imaging area of individual regions-of-interest (ROIs) that the CMOS image sensor chip can record at its full speed. Therefore, the whole field of view (FOV) of the image sensor was digitally divided into 16 (for human sperms) or 50 (for horse sperms) ROIs, which were sequentially recorded for continuous intervals of e.g., 0.7–7.0 seconds each. Further details of the human sperm imaging and tracking experiments can be found in Ref. 23. For horse samples, scanning over 50 such ROIs (with >5,000 lensfree holographic frames) and recording the 3D trajectories of >1,000 sperms took approximately 30 min for each semen sample. At the same time, the exposure time of the imaging system was also shortened to ~3 ms to avoid motion blur in recording the high-speed movement of horse sperms (which exhibit a typical instantaneous speed of e.g., ~150 μm/sec).
Reconstructing the 3D trajectories of sperms
For horse sperms, due to the high density of dead sperms and undissolved extender solute in the suspension liquid, each lensfree holographic frame was subtracted from a stationary image to remove the holograms of non-moving objects within the ROI. This stationary image was generated by averaging 100 consecutive lensfree frames that are nearest to the processing frame in the video sequence of the same ROI. These digital background cleaning steps were not needed and were not used for human sperm data.
The 3D trajectories of mobile sperms inside the FOV of the image sensor were then reconstructed following the procedures detailed in our previous work23. The vertical and oblique lensfree projections of each sperm head were digitally reconstructed on all the possible depth (i.e., z) planes individually. Once passed the morphological screening process, the centroid positions of both the vertical and the oblique head projections were calculated by their centers-of-gravity within their corresponding reconstructed amplitude images. The x and y coordinates of the sperms were taken directly from the centroid positions of the vertical head projections, while the z (depth) coordinates of the sperms were calculated by dividing the distance between their vertical and oblique projection centroids with the tangent of the oblique illumination angle in water. A space-time matrix containing the spatial and temporal coordinates of all the sperms within the observation volume was generated by repeating the same 3D localization procedures depicted above on all the holographic frames. Finally, the 3D trajectory of each sperm was constructed by linking the detected points across the recorded frames by a Brownian-statistics-based algorithm44. Note that the shapes of the sperm heads are assumed to be either spherical or ellipsoidal so that the orientation of the heads will not create a systematic error in the centroid-based position estimation. For tracking of sperms with deformed heads (see, e.g., images in References 45 and 46), processing techniques reported in Ref. 47 can potentially be used to minimize such errors and improve the reliability of our lensfree 3D tracking technique for deformed sperms.
Definitions of sperms' 3D dynamic swimming parameters
To quantify the 3D dynamics of sperm motion, we extracted a series of parameters from individual reconstructed sperm trajectories. All the parameters reported for horse sperms in this work were extracted from either 0.7 sec-long trajectories (~100 lensfree frames at 143 FPS) or track segments of such length that were digitally cropped from longer trajectories (e.g., ~4–7 sec long). For human sperms, however, these parameters were extracted from 1.1 sec-long tracks (~100 lensfree frames at 92 FPS). Similar to our previous work23, a digital “straightening” process was performed to compensate the curvature in sperm's forward motion before extracting these dynamical parameters. The definitions of parameters such as straight-line velocity (VSL), curvilinear velocity (VCL), linearity (LIN), amplitude of lateral head displacement (ALH) have been described in detail in our previous work23. Here we elaborate on the definition of the dynamical parameters that were newly introduced in this work:
Rate of twisting (RTW) represents the rotation speed (units: rad/sec) of the head beating plane for a sperm swimming in a ribbon pattern. It is defined as the angular frequency of the linear function that best fits the time evolution of the osculating plane angle for a track segment. The osculating plane angle on each position along the track segment is calculated by finding the most frequent angle of the lateral displacements occurring in the adjacent beating cycle, whose duration is defined by 2/BCF.
Twisting stability (TWS) is defined as the ratio between the accumulated angle change of the osculating plane and the averaged error to the best-fit linear function in the osculating plane angle. Both the angle change accumulation and the error averaging (by taking root mean square) were performed across the whole duration of each track segment. TWS represents how much a track segment is confined to a twisted ribbon (see, e.g., the magenta ribbon surfaces in Fig. 3 (b) and (e)). The value of TWS is reported in logarithm to the base 10. For example, a track segment with 10 radians of accumulated osculating plane angle change and 1 radians of mean linear-fit error would have a TWS of 1.
Digital classification of sperm trajectories
The 3D swimming patterns of sperms were classified based on the dynamic parameters defined in the previous section. Note that all the horse sperm trajectories with VCL smaller than 60 μm/sec and VSL smaller than 20 μm/sec are considered as immotile and are excluded from the reported statistics. The following are the specific criteria that we have used in this work to distinguish different categories of horse sperm trajectories:
Ribbon trajectory: TWS ≥ 1.2.
Hyperactivated trajectory: VCL ≥ 180 μm/sec and ALH ≥ 12 μm (following the definition used in Ref. 48).
Hyper-ribbon trajectory: A hyperactivated trajectory that also forms a ribbon (TWS ≥ 1.2).
Because human sperm trajectories were recorded at a lower frame rate (92 FPS instead of 143 FPS), the criteria for categorizing these trajectories for human sperms were modified as such:
Immotile trajectory: VCL < 30 μm/sec.
Ribbon trajectory: TWS ≥ 1.3.
Hyperactivated trajectory: VCL ≥ 150 μm/sec; LIN ≤ 0.5 and ALH ≥ 7 μm (following the definition used in Ref. 49).
The other criteria remained the same as the ones used for horse sperms.
Automated processing of 3D sperm trajectory data
Data processing procedures, including the reconstruction of lensfree holographic images, the localization of sperms' 3D centroids, the linking and resolution of sperms' 3D spatio-temporal trajectories and the classification of their 3D swimming patterns were all performed with a fully-automated custom-designed Matlab program. The typical computation time for automatic processing of e.g., ~5,000 lensfree images from a single semen sample is ~4 hours (using Matlab R2011a running on a PC with an eight-core Intel Core i7-930 2.80 GHz processor).
References
Maeda, K., Imae, Y., Shioi, J. I. & Oosawa, F. Effect of temperature on motility and chemotaxis of Escherichia coli. J. Bacteriol. 127, 1039–1046 (1976).
DiLuzio, W. R. et al. Escherichia coli swim on the right-hand side. Nature 435, 1271–1274 (2005).
Lauga, E., DiLuzio, W. R., Whitesides, G. M. & Stone, H. A. Swimming in Circles: Motion of Bacteria near Solid Boundaries. Biophysical Journal 90, 400–412 (2006).
Brokaw, C. J., Luck, D. J. & Huang, B. Analysis of the movement of Chlamydomonas flagella:"the function of the radial-spoke system is revealed by comparison of wild-type and mutant flagella". J Cell Biol 92, 722–732 (1982).
Rüffer, U. & Nultsch, W. High-speed cinematographic analysis of the movement of Chlamydomonas. Cell Motility 5, 251–263 (1985).
Phillips, D. M. Comparative analysis of mammalian sperm motility. J Cell Biol 53, 561–573 (1972).
Katz, D. & Overstreet, J. Sperm motility assessment by videomicrography. Fertil Steril 35, 188–193 (1981).
Mortimer, S. T. & Swan, M. A. Variable Kinematics of Capacitating Human Spermatozoa. Hum. Reprod. 10, 3178–3182 (1995).
Mortimer, S. T. CASA--practical aspects. J. Androl 21, 515–524 (2000).
Woolley, D. M. Motility of Spermatozoa at Surfaces. Reproduction 126, 259–270 (2003).
Friedrich, B. M., Riedel-Kruse, I. H., Howard, J. & Jülicher, F. High-precision tracking of sperm swimming fine structure provides strong test of resistive force theory. Journal of Experimental Biology 213, 1226–1234 (2010).
Berg, H. C. & Brown, D. A. Chemotaxis in Escherichia coli analysed by Three-dimensional Tracking. Nature 239, 500–504 (1972).
Crenshaw, H. C., Ciampaglio, C. N. & McHenry, M. Analysis of the Three-Dimensional Trajectories of Organisms: Estimates of Velocity, Curvature and Torsion from Positional Information. J Exp Biol 203, 961–982 (2000).
Xu, W., Jericho, M. H., Meinertzhagen, I. A. & Kreuzer, H. J. Digital in-line holography for biological applications. PNAS 98, 11301–11305 (2001).
Wu, M., Roberts, J. W., Kim, S., Koch, D. L. & DeLisa, M. P. Collective Bacterial Dynamics Revealed Using a Three-Dimensional Population-Scale Defocused Particle Tracking Technique. Appl. Environ. Microbiol. 72, 4987–4994 (2006).
Jericho, S. K., Garcia-Sucerquia, J., Xu, W., Jericho, M. H. & Kreuzer, H. J. Submersible digital in-line holographic microscope. Review of Scientific Instruments 77, 043706–10 (2006).
Sheng, J. et al. Digital Holographic Microscopy Reveals Prey-Induced Changes in Swimming Behavior of Predatory Dinoflagellates. PNAS 104, 17512–17517 (2007).
Corkidi, G., Taboada, B., Wood, C. D., Guerrero, A. & Darszon, A. Tracking sperm in three-dimensions. Biochem. Biophys. Res. Commun. 373, 125–129 (2008).
Drescher, K., Leptos, K. C. & Goldstein, R. E. How to track protists in three dimensions. Rev. Sci. Instrum. 80, 014301–7 (2009).
Langehanenberg, P. et al. Automated three-dimensional tracking of living cells by digital holographic microscopy. J. Biomed. Opt. 14, 014018 (2009).
Memmolo, P., Finizio, A., Paturzo, M., Miccio, L. & Ferraro, P. Twin-beams digital holography for 3D tracking and quantitative phase-contrast microscopy in microfluidics. Opt. Express 19, 25833–25842 (2011).
Restrepo, J. F. & Garcia-Sucerquia, J. Automatic three-dimensional tracking of particles with high-numerical-aperture digital lensless holographic microscopy. Opt. Lett. 37, 752–754 (2012).
Su, T.-W., Xue, L. & Ozcan, A. High-throughput lensfree 3D tracking of human sperms reveals rare statistics of helical trajectories. PNAS 109, 16018–16022 (2012).
Greenbaum, A. et al. Imaging without lenses: achievements and remaining challenges of wide-field on-chip microscopy. Nat Meth 9, 889–895 (2012).
Oda, R., Huc, I., Schmutz, M., Candau, S. J. & MacKintosh, F. C. Tuning bilayer twist using chiral counterions. Nature 399, 566–569 (1999).
Panyukov, S. & Rabin, Y. On the deformation of fluctuating chiral ribbons. Europhysics Letters (EPL) 57, 512–518 (2002).
Brizard, A., Ahmad, R. K. & Oda, R. Control of nano-micrometric twist and helical ribbon formation with gemini–oligoalanine via interpeptidic β-sheet structure formation. Chem. Commun. 2275–2277 (2007). 10.1039/B700959C.
Pashuck, E. T. & Stupp, S. I. Direct Observation of Morphological Tranformation from Twisted Ribbons into Helical Ribbons. J Am Chem Soc 132, 8819–8821 (2010).
Berg, H. C. How to Track Bacteria. Review of Scientific Instruments 42, 868–871 (1971).
Crenshaw, H. C. A New Look at Locomotion in Microorganisms: Rotating and Translating. Amer. Zool. 36, 608–618 (1996).
Colding, T. H. & Minicozzi, W. P. Shapes of embedded minimal surfaces. PNAS 103, 11106–11111 (2006).
Andersson, S., Hyde, S. T., Larsson, K. & Lidin, S. Minimal surfaces and structures: from inorganic and metal crystals to cell membranes and biopolymers. Chem. Rev. 88, 221–242 (1988).
Young, T. An Essay on the Cohesion of Fluids. Philosophical Transactions of the Royal Society of London 95, 65–87 (1805).
Mudanyali, O. et al. Wide-field optical detection of nanoparticles using on-chip microscopy and self-assembled nanolenses. Nat Photon 7, 247–254 (2013).
Hajduk, D. A. et al. The Gyroid: A New Equilibrium Morphology in Weakly Segregated Diblock Copolymers. Macromolecules 27, 4063–4075 (1994).
Falicov, A. & Cohen, F. E. A Surface of Minimum Area Metric for the Structural Comparison of Proteins. Journal of Molecular Biology 258, 871–892 (1996).
Elgeti, J., Kaupp, U. B. & Gompper, G. Hydrodynamics of Sperm Cells near Surfaces. Biophysical Journal 99, 1018–1026 (2010).
Crenshaw, H. C. & Edelstein-Keshet, L. Orientation by helical motion—II. Changing the direction of the axis of motion. Bulletin of Mathematical Biology 55, 213–230 (1993).
Crenshaw, H. C. Orientation by helical motion—III. Microorganisms can orient to stimuli by changing the direction of their rotational velocity. Bulletin of Mathematical Biology 55, 231–255 (1993).
Friedrich, B. M. & Jülicher, F. Chemotaxis of sperm cells. PNAS 104, 13256–13261 (2007).
Eisenbach, M. & Giojalas, L. C. Sperm guidance in mammals — an unpaved road to the egg. Nature Reviews Molecular Cell Biology 7, 276–285 (2006).
Kaupp, U. B., Kashikar, N. D. & Weyand, I. Mechanisms of Sperm Chemotaxis. Annual Review of Physiology 70, 93–117 (2008).
Bahat, A. & Eisenbach, M. Sperm thermotaxis. Molecular and Cellular Endocrinology 252, 115–119 (2006).
Crocker, J. C. & Grier, D. G. Methods of Digital Video Microscopy for Colloidal Studies. J. Colloid Interface Sci. 179, 298–310 (1996).
Brito, L. F. C. Evaluation of Stallion Sperm Morphology. Clinical Techniques in Equine Practice 6, 249–264 (2007).
Memmolo, P. et al. Identification of bovine sperm head for morphometry analysis in quantitative phase-contrast holographic microscopy. Opt. Express 19, 23215–23226 (2011).
Memmolo, P. et al. On the holographic 3D tracking of in vitro cells characterized by a highly-morphological change. Opt. Express 20, 28485–28493 (2012).
Rathi, R., Colenbrander, B., Bevers, M. M. & Gadella, B. M. Evaluation of In Vitro Capacitation of Stallion Spermatozoa. Biol Reprod 65, 462–470 (2001).
Mortimer, S. T., Swan, M. A. & Mortimer, D. Effect of seminal plasma on capacitation and hyperactivation in human spermatozoa. Hum. Reprod. 13, 2139–2146 (1998).
Acknowledgements
The authors thank Barbara Stewart (Veterinary Medical Teaching Hospital, University of California, Davis, CA, USA) and Stuart Meyers (Department of Anatomy Physiology and Cell Biology, University of California, Davis, CA, USA) for their valuable discussions and help on collecting horse semen specimens. A.O. acknowledges the support of the Army Research Office (ARO), ARO Young Investigator Award, National Science Foundation CAREER Award, the Office of Naval Research Young Investigator and PECASE Awards and the National Institutes of Health Director's New Innovator Award DP2OD006427 from the Office of The Director, National Institutes of Health.
Author information
Authors and Affiliations
Contributions
T.S. conducted the experiments and processed the resulting data. I.C., J.F. and K.H. contributed to experiments. E.M. contributed to the discussion section. T.S. and A.O. planned and executed the research. T.S., E.M. and A.O. wrote the manuscript. A.O. supervised the project.
Ethics declarations
Competing interests
A.O. is the co-founder of a start-up company that aims to commercialize lensfree microscopy tools.
Electronic supplementary material
Supplementary Information
Supplementary Movie S1
Supplementary Information
Supplementary Movie S2
Supplementary Information
Supplementary Movie S3
Supplementary Information
Supplementary Movie S4
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Su, TW., Choi, I., Feng, J. et al. Sperm Trajectories Form Chiral Ribbons. Sci Rep 3, 1664 (2013). https://doi.org/10.1038/srep01664
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01664
This article is cited by
-
A hybrid IMM-JPDAF algorithm for tracking multiple sperm targets and motility analysis
Neural Computing and Applications (2022)
-
Multifocal imaging for precise, label-free tracking of fast biological processes in 3D
Nature Communications (2021)
-
Advances in sperm analysis: techniques, discoveries and applications
Nature Reviews Urology (2021)
-
Self-organization and multi-line transport of human spermatozoa in rectangular microchannels due to cell-cell interactions
Scientific Reports (2020)
-
Bioinspired reorientation strategies for application in micro/nanorobotic control
Journal of Micro-Bio Robotics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.