Abstract
The actin crosslinking protein filamin A (FLNa) mediates mechanotransduction, a conversion of mechanical forces into cellular biochemical signals to regulate cell growth and survival. To provide more quantitative insight into this process, we report results using magnetic tweezers that relate mechanical force to conformational changes of FLNa immunoglobulin-like repeats (IgFLNa) 20–21, previously identified as a mechanosensing domain. We determined the force magnitudes required to unfold previously identified structural organizations of the β-strands in the two domains: IgFLNa 20 unfolds at ~15 pN and IgFLNa 21 unfolding requires significantly larger forces. Unfolded domain IgFLNa 20 can exist in two different conformational states, which lead to different refolding kinetics of the IgFLNa 20 and imply a significant impact on the reformation of the domain pair at reduced force values. We discuss the relevance of the findings to force bearing and mechanosensing functions of FLNa.
Similar content being viewed by others
Introduction
Actin filament (F-actin) networks provide mechanical stability of cells and their remodeling and contractility mediate mechanical events such as cell shape changes and movements. While chemical signals such as ATP hydrolysis and phosphorylation mediate remodeling and contraction of these networks, a reverse phenomenon, mechanotransduction, also occurs where mechanical forces elicit chemical changes that affect cell growth and survival1,2,3.
Filamins (FLNs), large actin-binding protein family, mediate these biological processes4,5,6. Since FLNs crosslink two actin filaments and have spring-like properties, they can potentially transmit and respond to mechanical forces applied to F-actin networks4,6,7. FLNs also bind and modulate multiple cellular components and disruption of these interactions results in impairment of binding partner's functions8,9,10. These findings suggest that FLN-partner interaction is at least in part regulated by mechanical force. Consistent with this hypothesis, externally imposed stress and internally generated contractile force to F-actin networks crosslinked by FLNa, which is the most abundant isoform among the vertebrate FLNs, regulate ligand binding to FLNa in vitro11.
FLNa is composed of two identical subunits, each containing an N-terminal actin-binding domain followed by 24 immunoglobulin-like (Ig) repeats (IgFLNa 1–24)12. The 24 Ig repeats can be divided into three segments: rod 1 (IgFLNa 1–15), rod 2 (IgFLNa 16–23) and Ig repeat 24 or dimerizing domain. Two short flexible hinge sequences separate these three segments. Repeats IgFLNa 1–15 in rod segment 1 form an extended linear structure without obvious inter-domain interactions. In contrast, rod segment 2 (IgFLNa 16–23) assumes a compact structure due to multiple inter-domain interactions. Domains 16–17, 18–19 and 20–21 form stable paired structures13,14. The compact rod 2 segment mediates most protein-protein interactions involving FLNa15.
Due to the unique domain-domain interactions among IgFLNa repeats 16–23, some binding interactions are considered auto-inhibitory. For example, the CD face of IgFLNa 21 provides the binding interface for the cytoplasmic tail of the β subunit of integrin, a key membrane adhesion molecule and migfilin, a promoter of integrin activation, while also interacting with the A-strand of IgFLNa 20 13,14. When IgFLNa repeats 20-21 are in their closed conformation, the A-strand of IgFLNa 20 blocks the CD face of IgFLNa 21, thus preventing integrin binding. Fully atomistic steered molecular dynamics (MD) simulations have inferred that mechanical force applied to this domain can expose the cryptic integrin binding site16, consistent with force-dependent binding of integrin peptide to FLNa in vitro11. Rognoni et al. recently reported that the disruption of the domain pair IgFLNa 20 and 21 occurs in the force range of 2–5 pN17. They estimated that domain pair disruption increases the binding affinity of β-integrin tail to IgFLNa 21 by ~17 fold. They also reported that at higher forces, the two domains mechanically unfold: IgFLNa 20 unfolding occurred at ~15 pN and that of IgFLNa 21 at > 30 pN.
Here we report independent confirmation of these quantitative unfolding results. In addition, we demonstrate that unfolded domain IgFLNa 20 has two distinct states with different degrees of stability that impact the refolding kinetics of IgFLNa 20 and therefore the reformation of the domain pair at lower forces. As the experimentally determined force of ~15 pN required to unfold domain IgFLNa 20 falls within a physiological range, this finding may provide further insights to the mechanosensensing function of the domain pair IgFLNa 20–21.
Results
We engineered and purified a recombinant protein construct consisting of IgFLNa 20–21: this pair of repeats was flanked by two identical “handles” comprised of IgFLNa 1–3 (Fig. 1a). As a control, we also generated a construct containing a single domain (IgFLNa 21) flanked by the same IgFLNa 1–3 “handles” (Fig. 1b). Both constructs featured a hexahistidine tag (His-tag) at the N-terminus and a biotinylated C-terminus. In the experiments, a tether was formed by attaching the His-tagged N-terminus to an NTA-copper coated coverslip surface, whereas the biotinylated C-terminus was attached to a 2.8-um-diameter streptavidin coated paramagnetic bead18,19. The tether was stretched by magnetic tweezers capable of generating >100 pN of force. The dimensional changes of the tether were recorded at a spatial resolution of ~2 nm and a sampling rate of ~100 Hz. The accuracy of force measurement had been confirmed by overstretching of short DNA20,21.
Schematics of protein constructs.
(a) Protein tether containing a pair of repeats IgFLNa 20-21 flanked by two handles consisting of repeats IgFLNa 1-3. The N-terminus is fused to a His-tag and is attached to a NTA-Cu coated cover glass surface, whereas the C-terminus is labeled with biotin and attached to a streptavidin-coated paramagnetic bead. (b) Control protein tether containing a single IgFLNa 21 domain spanned by the same handles.
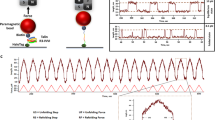
Fig. 2a shows the unfolding of four independent IgFLNa 20–21 tethers at a loading rate of ~2.1 pN/sec using the method described in previous published work18. For each of the IgFLNa 20–21 tethers, multiple stepwise unfolding signals were observed before the tether broke at high force magnitude (note that the extension coordinate is shifted for clarity). At this loading rate, unfolding of the domains was observed with forces ranging from ~10–120 pN. At low forces (~14–18 pN), the unfolding events occurred with a ~8 nm step size (Fig. 2a, right panel) and fluctuations between the unfolded state and folded states were occasionally observed (Fig. 2a, blue curve). At higher forces (30–120 pN), unfolding events resulted in larger step sizes of ~25 nm.
Unfolding of protein constructs.
(a) Four representative force-extension curves obtained from four different tethers containing IgFLNa 20–21 at a loading rate of ~2.1 pN/sec. The right panel shows zoomed-in curves at the force range of 12-20 pN where the unfolding of IgFLNa 20 occurs. The curves are shifted vertically in both panels for clarity. Raw data recorded at a sampling rate ~100 Hz are indicated by different colors for different tethers, which are smoothed by the adjacent-averaging method in a 0.1 second moving window (the white curves). (b) Extension step sizes and transition forces of all detected abrupt extension jumping events obtained from 29 protein tethers are shown as cross symbols. The histogram of the step sizes is shown on the right with three distribution peaks and the histogram of transition forces is shown on the top with a ~15 pN peak and widespread distribution to higher forces. (c) Four representative force-extension curves obtained from four different control tethers (indicated by different colors) at the same loading rate. The right panel shows zoomed-in curves at the force range of 12–20 pN, where no abrupt transitions are detected. The extensions of the different curves are shifted in both panels for clarity. Raw data and smoothed curves are obtained by the same method as with the IgFLNa 20–21 construct. (d) Extension step sizes and transition forces of all detected unfolding events obtained from 33 control tethers are shown as cross symbols. The histogram of the step sizes on the right panel shows a single peak and the histogram of transition forces shows widespread distribution lacking the peak at ~15 pN.
To validate our observations statistically, 29 independent protein tethers were stretched and a total of 139 unfolding or refolding events were analyzed. The results are summarized as scattered symbols of transition forces and step sizes in Fig. 2b. Three distinct data groups were revealed: two corresponding to unfolding events with a positive step size and one corresponding to refolding events with a negative step size. From these data, histograms of the force (top) and the step size (right) were generated. The force histogram reveals the highest probability distribution peak at ~15 pN, followed by unfolding forces spread over large range up to ~120 pN. The step size histogram reveals three distinct groups: ~25 nm unfolding steps under relatively large forces (>30 pN), ~8 nm unfolding steps under ~15 pN and around −8 nm refolding steps under ~15 pN. Based on worm-like chain model of polypeptide, the unfolded contour length can be obtained from unfolding force and step size (Supplementary Information: Fig. S1).
To identify the unfolding and refolding events specific to the IgFLNa 20–21 domain pair, we employed a control protein construct in which the IgFLNa 20–21 domain pair was replaced with a single IgFLNa 21 domain (Fig. 2c). As with the protein construct containing the IgFLNa 20–21 domain pair, we observed ~25 nm unfolding steps with widespread, large unfolding forces (most are greater than 30 pN). By contrast, the ~8 nm unfolding and refolding steps at ~15 pN were not observed with the control construct (Fig. 2d). These results indicated that the unfolding and refolding events resulting from application of ~15 pN force, with a step size of ~8 nm, are specific to the IgFLNa 20–21 domain pair.
The nature of the unfolding signals can be inferred by comparing our results with recent results obtained by Rognoni et al.17, which reported that i) domain pair disruption occurred at 2–5 pN, ii) unfolding of IgFLNa 20 occurred at ~15 pN and iii) unfolding of IgFLNa 21 occurred at >30 pN. Therefore, our observations are fully consistent with their results by assigning the unfolding event at ~15 pN with step size of ~8 nm to the unfolding of IgFLNa 20. The unfolding of IgFLNa 21, which was merged with the unfolding of other Ig domains (IgFLNa 1–3) in the two handles, occurred at higher force range with a typical step size of ~25 nm. Likely due to the limitations in sampling rate of our instrument, signal of the domain pair disruption in 2–5 pN was not observed in our experiments.
We obtained the results described above using a constant loading rate until the tether either unfolded completely or broke at high force. Since unfolding of IgFLNa20 occurs under physiologically relevant force range, we carefully examined mechanical properties of IgFLNa 20 subjected to low forces that are sufficient to unfold this domain while keeping IgFLNa21 intact. The curve depicted in black in Fig. 3a shows the extension change of a tether with a low loading rate of ~0.1 pN/second until the unfolding and refolding of IgFLNa 20 occurred (the spike in black curve), whereupon the force was kept constant at ~15.3 pN (the red colored curve). Rapid fluctuations with a rate of ~0.3 sec−1 between two distinct extension values (red curve left to the blue vertical dashed line) were observed, presumably corresponding to transitions between the folded state N and unfolded states of IgFLNa 20.
Unfolding of IgFLNa 20 under constant force.
(a) Time course of a protein tether containing IgFLNa 20–21 with different stretching forces. Black curve shows the extension recorded during increasing force at ~0.1 pN/sec loading rate. When the first stepwise extension fluctuation was observed, the magnets were stopped to maintain a constant force of ~15.3 pN (Red curve) for ~56 seconds. A stepwise fluctuation between two extension values occurred in the first ~18 seconds, followed by a stable longer extension in the following ~38 seconds. The blue dashed line separates the two kinetically distinct regions for clarity. Under subsequently reduced constant forces of 13.9 pN, 12.7 pN, 11.6 pN and 10.6 pN indicated by different colors, the unfolded IgFLNa 20 remained stable as no folding transition was observed. (b) Refolding and unfolding fluctuation at various constant forces from 16.1 – 19.5 pN obtained from another independent protein tether of IgFLNa 20–21. After unfolding at each force, the tether was refolded at low forces (~2 pN). At each force, similar stepwise fluctuation region and the stable longer extension region were observed, with a shorter lifetime of the stepwise fluctuation region at larger forces.
The rapid fluctuations lasted for only ~18 seconds, after which the protein tether stabilized in the unfolded state with longer extension (red curve right to the blue vertical dashed line). After ~30 seconds without observing a transition to the folded state with the shorter extension, the force was gradually decreased to ~10 pN. However, the refolding transition did not occur over the time scale of ~40 seconds at these smaller forces. These results strongly suggest the existence of two distinct unfolded states of IgFLNa 20 with different stabilities under ~15 pN force: a less stable unfolded state U1 experiencing rapid transitions with the folded state N with a lifetime of around three seconds and another more stable conformation U2 that remains unfolded even at decreased stretching forces of ~10 pN.
To see how the selection between the states U1 and U2 may be regulated by force, we studied the duration τ during which the rapid dynamic transitions between N and U1 states were observed before switching to the stable U2 state at constant forces in the range of 16.1 – 19.5 pN. At each force, after switching to the U2 state, the tether was refolded at low forces (~2 pN). The time traces shown in Fig. 3b clearly indicate shorter duration τ at larger forces. The reproducibility of such force-dependent duration τ was confirmed by experiments on multiple independent protein tethers, which also showed an overall trend of shorter duration τ at larger forces (Supplementary Information: Fig. S2).
The existence of two different unfolded states of IgFLNa 20 is also supported by the level of hysteresis in extension recorded between force-increase scan and the subsequent force-decrease scan. Fig. 4a show two force-extension curves recorded in two cycles of such force scans on the same tether over a force range around 2–18 pN at a loading rate of +0.5 pN/second in force-increase scan and −0.5 pN/second in force-decrease scan, respectively. In both cycles, the unfolding transition was observed in 15–18 pN (black lines in Fig. 4a) during the force-increase scans. However, the force-extension curves recorded in the force-decrease scans are different between the two cycles. In the first force-cycling scan, the refolding transition occurred at ~16 pN; whereas in the second force-cycling scan, refolding of IgFLNa 20 was not observed at a force of >8 pN. Such bimodal refolding transitions obtained on different tethers are also shown to demonstrate their reproducibility (Supplementary Information: Fig. S3). These results suggest that a simple two-state model cannot explain the transitions between the folded and unfolded states of IgFLNa 20. The observation of the two different refolding transitions associated with distinct refolding forces again indicates the existence of two unfolded states U1 and U2 with different degrees of stability.
Unfolding and refolding of IgFLNa 20 during force-cycling scans.
(a) Two successive force-extension curves obtained from another protein tether recorded at a constant loading rate of 0.5 pN/sec in the force-increase scan (black), followed by −0.5 pN/sec in the force-decrease scan (grey), over a force range up to ~18 pN. The curves are shifted in extension for clarity. In the first force-cycling scan, two-state fluctuations occurred at force ~17 pN and remained at a state with longer extension at the end of force-increase scan. In the force-decrease scan, unfolded IgFLNa 20 refolded at force ~16 pN indicated by a step-wise extension reduction. The similar transition forces of the unfolding in the force-increase scan and refolding in the force-decrease scan indicate a reversible transition between the two states with negligible hysteresis. In the second force-cycling scan on the same tether, similar observations of the transitions in 16–18 pN between unfolding and refolding of IgFLNa 20–21 were recorded in the force-increase scan. However, refolding of IgFLNa 20 was not observed in the same force range in the force-decrease scan. All shown curves were smoothed using a 0.1 second time window from the raw data. (b) Five force-cycling scans on another independent protein tether of IgFLNa 20–21 were also performed over a wider force range up to 22 pN by changing the distance between the magnets and bead d (Supplementary Information: Magnetic tweezers setup) at a constant speed of 20 μm/sec. Unlike the constant loading rate force-scanning, this constant speed force-scanning procedure has an advantage of allowing protein refolding at low force for a longer time (Supplementary Information: Fig. S4). In all the force-decrease scans, refolding steps under the forces greater than 10 pN were not observed. Two of the four force-increase scans (excluding the first scan for the initial native protein tether) showed unfolding signals in 13–18 pN, indicating refolding probability of ~50% at low forces in our force-scanning procedure.
The force-dependent selection between the states U1 and U2 was also investigated by force-cycling scanning experiments. Fig. 4a and Fig. S3 have shown that scanning up to ~18 pN results in nearly equal probability of the selection between U1 and U2. Fig. 4b shows five successive force-cycling scans obtained from another independent experiment over a wider force range up to ~22 pN. In all the five force-decrease scans, rapid refolding and unfolding transitions were not observed, indicating only the U2 state was selected at the higher force of ~22 pN during the force-increase scans. After the U2 state was selected, even during force-decrease scans to a very low force range of ~2.4 to 4 pN under which the domain pair IgFLNa 20–21 may form17, the stable unfolded IgFLNa 20 conformation U2 still had significant probability (~50%) of being unable to refold at a time scale of 50 seconds (Fig. 4b and Supplementary Information: Fig. S4).
Fig. 5 depicts a hypothetical three-state model to describe the unfolding and refolding transitions of IgFLNa 20. The model includes a folded state N, an intermediate unfolded state U1 and a more stable unfolded state U2. The two unfolded states likely exhibit different conformations, but the difference in extension is too small to be resolved by the magnetic tweezers used in our experiments. In the absence of tension, the folded state N is the most stable. As force increases, the free energy landscape tilts, and, at ~15 pN, the state N and U1 have comparable free energies and rapid transitions between the two states occur (assuming a small energy barrier between the two states). When the tether is held for sufficiently long time under force >15 pN, a transition from the intermediate U1 state to the more stable U2 state occurs. A larger energy barrier separating the U1 and U2 states may account for the slower transition rate between U1 and U2. As U2 is more stable than U1, the U2-to-N transition will not occur under the same force of ~15 pN. The possible physical pictures of the two unfolded states U1 and U2 will be discussed in the Discussion section.
Hypothetical model of two different unfolded IgFLNa 20 conformations.
(a) A folded state N and two hypothesized unfolded states: an intermediate state U1 and a more stable state U2. The transition between states N and U1 is reversible under force of ~15 pN, while a subsequent transition from U1 to a more stable conformation U2 is irreversible. (b) Hypothesized free energy landscape of IgFLNa 20. At zero force, the folded state N is the most stable. Under ~15 pN of force, states N and U1 have similar free energies and are separated with small energy barrier, resulting reversible transition between the two states. In contrast, transition from the U1 state to the more stable U2 state is irreversible, requiring overcoming a large energy barrier.
Discussion
Our results demonstrated that unfolded domain of IgFLNa 20 can exist in two different conformational states U1 and U2, which lead to different refolding kinetics of the IgFLNa 20. Though the two unfolded states U1 and U2 have similar extensions, they have different stability and kinetic behavior. We discuss two possibilities that may explain our observations. One possibility is that unfolded chains might associate with each other in the U2 state, leading to a larger energy barrier for refolding. However, the possibility of this event in single-molecule stretching experiments is small because 1) there are no free proteins in solution and 2) the self-association of the unfolded domains is unfavorable under large tension. We instead propose a second possibility that the folding from the unfolded state to the native state N requires a nucleation state U1 which contains a tiny folded “seed” that adsorbs a small peptide with length of l. The U2 state then represents a state where the “seed” is unfolded. In this picture, l should be shorter than our instrumentation spatial resolution of ~2 nm since we did not directly observe it in experiments. Although the exact physical picture of the nucleated “seed” is not clear, we consider a possibility that the seed is a small but stable β-hairpin structure that requires looping of a short random coiled peptide chain. In this picture, the “nucleation” rate under force f is roughly proportional to a factor Exp(-fl/kBT), where kB is the Boltzmann constant and T is the absolute temperature. At a force ~15 pN where the U1 to U2 transition was observed and with l ~2 nm, this factor is small (<10−3), which can explain why refolding from U2 is much slower than refolding from U1 states. A possible analogy is the zipping of an unzipped DNA hairpin with a random coiled single-stranded end-loop. By choosing the sequences of the DNA hairpin and the size of the end-loop, we were able to produce a free energy profile similar to that sketched in Fig. 5b (Supplementary Information: Fig. S5).
These findings have biological implications for the mechanosensing function of FLNa. As the ~15 pN force required to unfold IgFLNa 20 approximates the force generated by several myosin motors (12), unfolding of IgFLNa 20 may occur in vivo. We showed that the mechanically unfolded domain IgFLNa 20 could have two different conformations with different stabilities. The less stable unfolded IgFLNa 20 can reversibly refold at force ~15 pN, while the more stable unfolded IgFLNa 20 exhibits a much slower refolding kinetics even at low force of smaller than 4 pN. According to the results reported by Rognoni et al.17, disruption of the domain pair IgFLNa 20–21 occurred at a similar force range of 2–5 pN. Therefore, our result implies that the more stable unfolded conformation of IgFLNa 20 will significantly slow down the reformation of the domain pair at low forces, which may further enhance the binding of β-integrin tail to IgFLNa 21 by exposing the CD-face of IgFLNa 21 for a longer duration. Compared to Rognoni et al.’s results that indicate an equilibrium mechanosensing mechanism over a force range of 2–5 pN, our results complementarily suggest a non-equilibrium mechanosensing mechanism and expand the force sensing range to ~15 pN.
Methods
A protein construct of IgFLNa repeats composed of IgFLNa 20–21 flanked by two handles consisting of IgFLNa 1–3 was designed. In addition to separating the domain pair IgFLNa 20–21 from the surfaces of coverslip and paramagnetic bead, the handles also provide signature of regular unfolding steps to avoid non-specific tether (which lacks the regular unfolding step) or multiple protein tethers (which deflects the bead position in the focal plane) formed between the bead and the surface18. A protein construct with the IgFLNa 21 flanked by the same handles was expressed as a control to identify signals specific to IgFLNa 20–21. Both protein constructs were labeled with His-tag at N-terminal and Avi-tag at C-terminal which was biotinylated. The DNA constructs were transfected into sf-9 insect cells and recombinant proteins were purified as previously described18,20.
A magnetic tweezers setup was developed and used to apply force to the protein constructs20 (Supplementary Information: Magnetic tweezers setup). The N-terminal His-tag of the protein construct was bound to the NTA-copper coated surface of glass coverslip, whereas the C-terminal biotin was bound to a streptavidin-coated paramagnetic bead (Dynal M-270 bead, Invitrogen). Using permanent magnets placed above the sample, stretching forces were applied through the paramagnetic bead and forces were controlled by changing the distance between the sample and the magnets. The extension of the protein construct was measured by comparing the image of the paramagnetic bead with pre-stored images of the same bead at a series of different focal planes22. Drift was canceled by a reference polystyrene bead stuck on the coverslip surface.
When the magnet's position is fixed, the force applied to the protein is a constant. With the pre-calibrated force as a function of the distance d between the permanent magnets and the sample, the magnets can approach the sample continuously to increase force exponentially or linearly with time18.
References
Brown, A. E. & Discher, D. E. Conformational changes and signaling in cell and matrix physics. Current biology : CB 19, R781–789 (2009).
Moore, S. W., Roca-Cusachs, P. & Sheetz, M. P. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Developmental cell 19, 194–206 (2010).
Schwartz, M. A. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol 2, a005066 (2010).
Stossel, T. P. et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol 2, 138–145 (2001).
Janmey, P. A. & McCulloch, C. A. Cell mechanics: integrating cell responses to mechanical stimuli. Annual review of biomedical engineering 9, 1–34 (2007).
Razinia, Z., Makela, T., Ylanne, J. & Calderwood, D. A. Filamins in mechanosensing and signaling. Annual review of biophysics 41, 227–246 (2012).
Furuike, S., Ito, T. & Yamazaki, M. Mechanical unfolding of single filamin A (ABP-280) molecules detected by atomic force microscopy. FEBS Lett 498, 72–75 (2001).
Gawecka, J. E., Griffiths, G. S., Ek-Rylander, B., Ramos, J. W. & Matter, M. L. R-Ras regulates migration through an interaction with filamin A in melanoma cells. PLoS ONE 5, e11269 (2010).
D'Addario, M., Arora, P. D., Ellen, R. P. & McCulloch, C. A. Interaction of p38 and Sp1 in a mechanical force-induced, beta 1 integrin-mediated transcriptional circuit that regulates the actin-binding protein filamin-A. The Journal of biological chemistry 277, 47541–47550 (2002).
Nakamura, F., Stossel, T. P. & Hartwig, J. H. The filamins: Organizers of cell structure and function. Cell Adh Migr 5 (2011).
Ehrlicher, A. J., Nakamura, F., Hartwig, J. H., Weitz, D. A. & Stossel, T. P. Mechanical strain in actin networks regulates FilGAP and integrin binding to filamin A. Nature 478, 260–263 (2011).
Nakamura, F., Osborn, T. M., Hartemink, C. A., Hartwig, J. H. & Stossel, T. P. Structural basis of filamin A functions. J Cell Biol 179, 1011–1025 (2007).
Heikkinen, O. K. et al. Atomic Structures of two novel immunoglobulin-like domain pairs in the actin cross-linking protein filamin. Journal of Biological Chemistry 284, 25450–25458 (2009).
Lad, Y. et al. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J 26, 3993–4004 (2007).
Nakamura, F., Stossel, T. P. & Hartwig, J. H. The filamins: Organizers of cell structure and function. Cell Adh Migr 5, 160 (2011).
Pentikainen, U. & Ylanne, J. The regulation mechanism for the auto-inhibition of binding of human filamin A to integrin. J. Mol. Biol. 393, 644–657 (2009).
Rognoni, L., Stigler, J., Pelz, B., Ylanne, J. & Rief, M. Dynamic force sensing of filamin revealed in single-molecule experiments. Proc Natl Acad Sci U S A 109, 19679–19684 (2012).
Chen, H. et al. Differential mechanical stability of filamin A rod segments. Biophys J 101, 1231–1237 (2011).
del Rio, A. et al. Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 (2009).
Chen, H. et al. Improved high-force magnetic tweezers for stretching and refolding of proteins and short DNA. Biophys J 100, 517–523 (2011).
Fu, H. et al. Transition dynamics and selection of the distinct S-DNA and strand unpeeling modes of double helix overstretching. Nucleic Acids Res 39, 3473–3481 (2011).
Gosse, C. & Croquette, V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys J 82, 3314–3329 (2002).
Acknowledgements
This research was supported by National Research Foundation of Singapore through the Mechanobiology Institute at National University of Singapore (JY) as well as the HUSEC Seed Fund for Interdisciplinary Science (FN and TPS) and National Institute of Health grant HL19749 (TPS).
Author information
Authors and Affiliations
Contributions
H.C. and S.C. performed the experiments. M.P.S., T.P.S., F.N. and J.Y. conceived the research. H.C., F.N. and J.Y. designed the experiments and interpreted the data. H.C., T.P.S., F.N. and J.Y. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Chen, H., Chandrasekar, S., Sheetz, M. et al. Mechanical perturbation of filamin A immunoglobulin repeats 20-21 reveals potential non-equilibrium mechanochemical partner binding function. Sci Rep 3, 1642 (2013). https://doi.org/10.1038/srep01642
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01642
This article is cited by
-
The role of single-protein elasticity in mechanobiology
Nature Reviews Materials (2022)
-
Prune belly syndrome in surviving males can be caused by Hemizygous missense mutations in the X-linked Filamin A gene
BMC Medical Genetics (2020)
-
Mechanical unfolding of spectrin reveals a super-exponential dependence of unfolding rate on force
Scientific Reports (2019)
-
When signalling goes wrong: pathogenic variants in structural and signalling proteins causing cardiomyopathies
Journal of Muscle Research and Cell Motility (2017)
-
Talin Dependent Mechanosensitivity of Cell Focal Adhesions
Cellular and Molecular Bioengineering (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.