Abstract
The red flour beetle, Tribolium castaneum, has been widely used as a laboratory model for analyzing gene function. In this study, we established a novel cell line (Tc81) from T. castaneum embryos and validated the utility of this cell line by analyzing the juvenile hormone (JH) signaling pathway. In Tc81 cells, the Krüppel homolog 1 gene (Kr-h1), which is a JH-dependent repressor of insect metamorphosis, was rapidly induced by subnanomolar levels of JHs. Bioinformatics analysis and reporter assays identified 2 JH response elements (kJHREs) located in the region upstream of the transcription start site and in the first intron of Kr-h1. Furthermore, methoprene tolerant (Met) and steroid receptor co-activator (SRC) RNAi reduced JH-dependent induction of Kr-h1 transcripts and kJHRE-reporter activities. Thus, this novel Tc81 cell line is useful for the elucidation of JH signaling and is a promising tool for the functional analysis of genes by RNAi and reporter assays.
Similar content being viewed by others
Introduction
The red flour beetle Tribolium castaneum is a serious pest often found in stored agricultural products and is widely used as a laboratory model1,2. T. castaneum has many advantages as an experimental organism. For example, it has a relatively short life cycle (about 4 weeks from embryo to adult)3 and is easy to maintain. The elucidation of complete annotated genomic sequences4, transposon-based genetic transformation5 and systemic RNA interference (RNAi)3 has facilitated the analysis of gene function in this model. In addition, unlike the fruit fly Drosophila melanogaster, which has a highly derived developmental pattern, T. castaneum development follows a typical process that can be considered representative of primitive holometabolous insects3,4. These features have made this species an indispensable model insect for research in evolutional and developmental biology, as well as the study of pest management.
One of a few disadvantages of this experimental insect is the shortage of cell lines derived from the organism. Although over 500 cell lines have been established from various tissue sources of many insect species, cell lines of T. castaneum have not yet been established, except for one such cell line reported very recently by Goodman et al.6. Some of these insect cell lines have been used as research tools to elucidate functions and regulatory mechanisms of genes involved in various biological phenomena7. In particular, they are useful for the functional analysis of genes involved in complex signaling pathways, where the functions of individual genes would be too difficult to determine using whole organisms. In addition, these cell lines are valuable for the development of efficient screening systems to discover new drugs, including insecticides7. Therefore, the establishment of new T. castaneum cell lines will undoubtedly enhance the value of this model insect.
Juvenile hormones (JHs) comprise a group of sesquiterpenoids that regulate a wide array of developmental and physiological events in insects, such as metamorphosis, reproduction, diapause and polyphenism8,9. JH is known as a “status quo” hormone and is necessary for maintaining larval nature during molting and for repressing metamorphosis10. Although the molecular mechanisms facilitating the antimetamorphic action of JH have long been a mystery11, recent breakthroughs in the study of T. castaneum have largely expanded our knowledge of these processes12.
Methoprene-tolerant (Met) and Krüppel homolog 1 (Kr-h1) have critical roles in the JH signaling cascade during metamorphosis12. Met is a basic helix-loop-helix Per-ARNT-Sim (bHLH–PAS) transcription factor that was initially identified from a D. melanogaster mutant as a resistance gene to the JH agonist methoprene13,14. Kr-h1 is a C2H2 zinc-finger-type transcription factor that was initially identified as a modulator of the prepupal ecdysone response in D. melanogaster15; it was later found to be induced by JH16. RNAi analysis of Met (TcMet) in the larvae and pupae of T. castaneum revealed that JH carries out its antimetamorphic action via TcMet17,18. Additional RNAi analyses in T. castaneum revealed that JH induces Kr-h1 (TcKr-h1), a metamorphosis inhibitor, via TcMet, thereby mediating the antimetamorphic activity of JH19. Importantly, this JH-Met-Kr-h1 cascade is conserved in the larval-pupal transition in holometabolous insects and the nymphal-adult transition in hemimetabolous insects19,20,21.
Very recently, Met was found to form a functional JH receptor with another bHLH-PAS transcription factor, steroid receptor co-activator (SRC, also named FISC and Taiman), based on studies in D. melanogaster, Aedes aegyptii and T. castaneum12. Met proteins bound to JH with high affinity22,23, heterodimerized with SRC in a JH-dependent manner and stimulated the transcription of JH-inducible genes, such as Kr-h1 and early trypsin (ET)24,25. Furthermore, using a cell line derived from A. aegyptii, Li et al.24 identified a JH response element (JHRE) that interacts with Met and SRC in the promoter region of the ET gene.
Using a cell line derived from Bombyx mori, we previously identified a different JHRE (kJHRE) in the promoter of the Kr-h1 (BmKr-h1) gene and revealed that the JH/Met/SRC complex uses kJHRE to activate BmKr-h1 transcription26. Intriguingly, sequences similar to the kJHRE are also present in the promoter of Kr-h1s in T. castaneum and other insect species26, suggesting their conserved and relevant roles in JH signaling. However, the function of these kJHRE-like sequences in T. castaneum and other insects remains to be characterized.
In this study, we established a novel cell line (Tc81) from T. castaneum embryos and used this cell line for molecular analysis of the JH signaling pathway. Using a combination of RNAi and reporter gene assays in Tc81 cells, we analyzed the functions of Met, SRC and kJHREs in the JH-dependent induction of Kr-h1 in T. castaneum. This is the first report to describe the establishment of a T. castaneum-derived cell line that is applicable for the RNAi-based functional analysis of various genes.
Results
Characterization of Tc81 cells
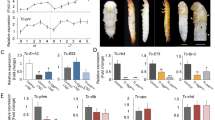
Cells derived from T. castaneum embryos (Tc81) were suspended throughout the majority of the culture medium, with vesicles forming and cells occasionally adhering to the bottom of the culture flask (Fig. 1A). The origin of the Tc81 cells was confirmed by the sequences of 3 representative genes in the genomic DNA of the cells, which perfectly matched with the previously published sequences of the respective T. castaneum genes6 (Supplementary Fig. 1A online). DAPI staining of the nuclei of Tc81 cells suggested that each lattice in the Fig. 1A inset represented a cell (Fig. 1B). The majority of Tc81 cells contained 20 chromosomes/cell, which was double the standard number of chromosomes for in vivo haploid T. cataneum cells (10 chromosomes)6, indicating that Tc81 cells are mainly diploid (Supplementary Fig. 1B online). The size and shape of individual Tc81 cells were uniform, measuring about 10 μm in diameter, while the shape of vesicles was variable, with sizes ranging from about 30 to 300 μm in length (Fig. 1A). The vesicles were collected by centrifugation and dispersed into fresh medium by gentle pipetting (Fig. 1C). After this manipulation, most vesicles temporarily withered, but supple vesicles were regenerated within 2 days. After transfer to fresh medium, cell numbers decreased slightly, but started to increase again after day 2 (Fig. 1D).
Morphology and growth capacity of Tc81 cells.
(A) Phase contrast micrograph of Tc81 cells forming vesicles. (B) Fluorescence micrograph of Tc81 cells stained with DAPI. (C) Phase contrast micrograph of vesicles disrupted by centrifugation and pipetting. (D) Growth curves of Tc81 cells after 120 passages. The cultures were maintained at 25°C. Mean ± SD (n = 4).
Efficiency of soaking Tc81 cells in RNAi
To evaluate the efficiency of RNAi in Tc81 cells, we selected the JH receptor TcMet as a target gene and MalE of Escherichia coli as a control. Cells were soaked with medium containing one of these dsRNAs and the expression of TcMet was analyzed by qPCR. In cells treated with TcMet dsRNA, TcMet transcripts declined rapidly to about half of baseline levels by day 1 and to about one-third of baseline levels by day 2, maintaining this reduced expression until day 8 (Fig. 2A). In contrast, in cells soaked with MalE dsRNA, TcMet transcript expression was similar to that in untreated cells and showed little decline throughout the experiment (Fig. 2A). These results suggested that the decline in TcMet transcript expression observed in the cells soaked with TcMet dsRNA was a target-specific effect. No further suppression of TcMet transcripts was achieved by additional dsRNA treatment, indicating that repeated treatment was not necessary to enhance RNAi efficiency, at least until day 8. Fig. 2B shows the relationship between the concentration of dsRNA and the suppression of target transcripts. Expression of TcMet transcripts was decreased by treatment with TcMet dsRNA in a concentration-dependent manner (0.0005–0.5 ng/μL). Virtually no further suppression was observed at higher concentrations (0.5–500 ng/μL). A near maximal RNAi effect was obtained at a concentration of 0.05 ng/μL dsRNA (Fig. 2B). Treatment with dsMalE dsRNA showed no significant effects on TcMet transcript expression at any concentration tested (0.0005–500 ng/μL; Fig. 2B).
Effects of soaking Tc81 cells in TcMet dsRNA.
TcMet transcript levels were determined by qPCR (mean ± SD, n = 3). (A) Cells were soaked with TcMet (dsMet) or MalE (dsMalE, control) dsRNA (50 ng/μL) and temporal changes in TcMet transcript levels were measured (dsMalE and dsTcMet). Some cells were retreated with dsRNA (50 ng/μL) 4 days after the first treatment (+dsMalE and +dsTcMet). (B) Cells were treated with different concentrations of dsRNA for MalE or TcMet and the expression levels of TcMet were determined 60 h after treatment.
Effects of JH and its analog on the expression of TcKr-h1 in Tc81 cells
The effects of JH and its analog (methoprene; JHA) on the expression of TcKr-h1 in Tc81 cells were analyzed by qPCR. TcKr-h1 transcript expression was marginal before JHA treatment, but increased significantly within 0.5 h after treatment, reaching a peak (1.5 × 102-fold increase in expression compared to baseline levels) at 1 h after treatment (Fig. 3A). The expression decreased to about half of the maximum level by 2 h and was maintained at this level until 24 h after treatment (Fig. 3A). JH I, JH III and JHA induced TcKr-h1 transcript expression dose-dependently in Tc81 cells; the 50% effective concentrations (EC50s) were 4.7 × 10−11, 2.7 × 10−11 and 1.8 × 10−9 M, respectively (Fig. 3B). Farnesoic acid (FA) stimulated weak induction of TcKr-h1 transcripts, but only at a high concentration (1 μM; Fig. 3B).
Induction of TcKr-h1 transcripts by JH in Tc81 cells.
Transcript levels of TcKr-h1 were determined by qPCR (mean ± SD, n = 3). (A) Cells were treated with 10 μM methoprene (JHA) and temporal changes in TcKr-h1 expression were monitored. (B) Cells were treated with different concentrations of JH I, JH III, JHA, or farnesoic acid (FA) and the expression levels of TcKr-h1 were determined 2 h after treatment.
Effects of TcMet and TcSRC RNAi on the JH-dependent induction of TcKr-h1 in Tc81 cells
Next, we analyzed the functions of TcMet and SRC (TcSRC) in T. castaneum in the JH-dependent induction of TcKr-h1 by RNAi in our newly established Tc81 cell line. Since only a partial sequence for TcSRC has been reported in public databases and the literature23,25, we cloned the full-length cDNA of TcSRC (AB762694) by RACE. The open reading frame (ORF) encoded a predicted protein with 1,344 amino acid residues. Four domains, i.e., bHLH, PASA, LXXLL and PolyQ, of TcSRC showed high homology with the SRCs of other insect species (Supplementary Fig. 2 online).
In Tc81 cells soaked with dsRNA for TcMet or TcSRC, the transcript levels of the respective target genes were significantly reduced compared to untreated cells or cells treated with MalE dsRNA (Fig. 4A and B). The application of JHA did not significantly affect the expression of TcMet or TcSRC in either control or test cells (Fig. 4A and B). In contrast, TcKr-h1 transcript expression in Tc81 cells was low before JHA treatment, but dramatically increased following JHA treatment. However, this induction by JHA was significantly suppressed in cells treated with TcMet or TcSRC dsRNAs (Fig. 4C). These results confirmed that both TcMet and TcSRC were involved in the signaling pathway mediating JH-induced TcKr-h1 expression.
Effects of RNAi-mediated knockdown of TcMet and TcSRC on the JH-dependent induction of TcKr-h1 in Tc81 cells.
Cells were soaked in 50 ng/μL dsRNA for MalE, TcMet, or TcSRC for 60 h. Control cells (−) were not treated with dsRNA. Cells were then incubated in media containing 10 μM JHA (JHA+) or JHA-free media (JHA-) for 2 h and TcMet (A), TcSRC (B) and TcKr-h1 (C) transcript levels were determined by qPCR (mean ± SD, n = 3). Data were analyzed using Student's t-tests (***P < 0.001; **P < 0.01; *P < 0.05; not indicated, P > 0.05).
Identification and functional characterization of kJHREc in the TcKr-h1 gene
Next, we searched for kJHREs in Kr-h1 genes from 4 insect species. This search yielded candidate sequences in the first intron in addition to ones previously identified upstream of the first exon (Fig. 5A). These 11-bp sequences (GCCTCCACGTG) found in the first intron of Kr-h1 in B. mori (2 sequences), T. castaneum (1 sequence) and A. mellifera (1 sequence) matched the original kJHREc identified upstream of BmKr-h1 perfectly (Fig. 5B, top). Two putative kJHREcs, each having 1- or 2-nucleotide substitutes in the 5′ region, were also found in the first intron of the Kr-h1 gene in D. melanogaster (Fig. 5A, B). All of these sequences contained an identical canonical E-box sequence (CACGTG).
Prediction and functional determination of JHREs of TcKr-h1.
(A) Schematic representation of core JHRE (kJHREc)-like sequences present upstream and within the first intron of Kr-h1 genes from 4 insect species. Tc, T. castaneum; Bm, B. mori; Am, A. mellifera; Dm, D. melanogaster. White boxes, black boxes and arrows represent the 5′-UTRs, ORFs and transcription start sites, respectively. The numbers indicate distances from the transcription start site. Red ellipses represent the authentic kJHREc (-2079/-2068) in B. mori26 and its homologs. The asterisk shows a gap in the genomic sequence. (B) Alignment of the putative kJHREc sequences is shown in (A). (C) Functional characterization of putative kJHREcs of TcKr-h1. Tc81 cells were cotransfected with reporter plasmids that express firefly luciferase under the regulation of indicated regions and a reference reporter plasmid carrying Renilla luciferase. Red Xs indicate mutations in putative kJHREc sequences (see Materials and Methods for details). Cells were treated with 10 μM JHA for 24 h and reporter activity was measured using a dual-luciferase reporter assay system. The activity of the firefly luciferase reporter was normalized against that of the Renilla luciferase reporter in the same samples. Data represent means ± SD (n = 3). Means with the same letter are not significantly different (Tukey–Kramer test, P < 0.05).
To characterize the function of the putative kJHREcs found in the TcKr-h1 gene in the context of JH stimulation, we carried out reporter assays in Tc81 cells. We tested reporter constructs carrying the upstream and first intron region (−477 to +1883), the upstream region alone (−477 to +106) and the first intron region alone (+106 to +1883). All 3 constructs showed 8- to 12-fold increases in luciferase reporter activity in the presence of JHA (Fig. 5C). When a mutation (CGCCTCCACGTG to TTTAAATTTAAA) was introduced in one or both of the kJHREcs in the −477 to +1883 region, reporter activity in the presence of JHA decreased by 51%–63% with single mutations and by 73% with double mutations, as compared to the wild-type reporter (Fig. 5C). When kJHREcs were mutated in the reporter harboring the upstream region alone (−477 to +106) or the first intron region alone (+106 to +1883), JHA-dependent stimulation was completely abolished (Fig. 5C). These results indicated that the 2 kJHREcs located upstream of the transcription start site and first intron of TcKr-h1 contributed independently and additively to the JH-dependent induction of TcKr-h1.
Effects of TcMet and TcSRC RNAi on kJHRE reporter activity in Tc81 cells
To examine the interactions of TcMet, TcSRC and kJHRE in the JH-dependent induction of TcKr-h1, we further performed reporter assays in Tc81 cells using a kJHRE reporter vector (−477 to +1883) in combination with TcMet or TcSRC RNAi. The reporter activities induced by JHA treatment were decreased by about 75% in cells treated with TcMet or TcSRC dsRNA, as compared to untreated control cells or cells treated with MalE dsRNA (Fig. 6). This result was consistent with the effects of TcMet and TcSRC RNAi on the JH-dependent induction of TcKr-h1 transcripts (Fig. 4C), indicating that TcMet and TcSRC induced the transcription of the TcKr-h1 gene via interaction with the kJHRE in the presence of JH.
Effects of TcMet and TcSRC RNAi and JHA treatment on reporter activity in Tc81 cells.
Tc81 cells were cotransfected with a reporter plasmid carrying a kJHRE region (−477 to +1883, pGL4.14) conjugated to firefly luciferase, a reference reporter plasmid carrying Renilla luciferase and the indicated dsRNAs. The cells were treated with 10 μM methoprene (JHA) for 24 h and reporter activities were measured using a dual-luciferase reporter assay system. The activity of the firefly luciferase reporter was normalized to that of the Renilla luciferase reporter in the same samples. Data represent means ± SD (n = 3). Data were analyzed using Student's t-tests (***P < 0.001; **P < 0.01; *P < 0.05; not indicated, P > 0.05).
Analysis of the interactions between TcMet, TcSRC and kJHRE in mammalian cells
Next, we examined the JH-dependent interaction of TcMet and TcSRC using full ORFs of these proteins in mammalian 2-hybrid reporter assays. As expected, no JHA-dependent increase in UAS reporter activity was observed when HEK293 cells were transfected with Gal4DBD-TcMet, Gal4DBD-TcSRC, VP16AD-TcMet, or VP16AD-TcSRC (Fig. 7A). In contrast, when cells were cotransfected with Gal4DBD-TcMet:VP16AD-TcSRC or Gal4DBD-TcSRC:VP16AD-TcMet, UAS reporter activity was significantly increased in the presence of JHA (Fig. 7A). These results suggested that the full ORFs of TcMet and TcSRC proteins interacted with each other in a JH-dependent manner in HEK293 cells.
Functional analysis of TcMet, TcSRC and kJHRE in mammalian HEK293 cells.
(A) Cells were cotransfected with a UAS reporter plasmid carrying firefly luciferase, a reference reporter plasmid (pRL-TK) carrying Renilla luciferase and an expression plasmid carrying GAL4DBD or VP16AD fused with TcMet or TcSRC. Cells were then treated with 0.1 μM JH III for 24 h. (B) Cells were cotransfected with a kJHRE-reporter vector (−477 to +1883, pGL4.14) and a plasmid expressing the full ORF of TcMet or TcSRC. Cells were then treated with 0.1 μM JH III for 24 h. Xs indicate mutations in the kJHREc. (A, B) Reporter activity was examined using a dual-luciferase reporter assay system. Data represent means ± SD (n = 3). Data were analyzed using Student's t-tests (***P < 0.001; **P < 0.01; *P < 0.05; not indicated, P > 0.05). (C) Schematic representation of the interactions of kJHRE, TcMet and TcSRC in the JH-dependent transcription of TcKr-h1. X represents an additional cofactor.
Finally, we examined whether the kJHRE reporter (−477 to +1883) containing the 2 kJHREc sequences could be induced by JH in HEK293 cells and Drosophila S2 cells in which full-length TcMet and TcSRC proteins were heterologously expressed.
The wild-type reporter showed no JH-dependent induction in HEK293 cells in which neither TcMet nor TcSRC were expressed (Fig. 7B). Similarly, when only one construct (i.e., TcMet or TcSCR) was expressed, the wild-type reporter again exhibited no JH-dependent induction. In contrast, weak but significant JH-dependent induction was observed in cells in which both TcMet and TcSRC were expressed. Similar levels of JH-dependent induction were observed when only the upstream kJHRE was mutated in the reporter vector; this effect was abolished when the kJHRE in the first intron was mutated or when both kJHREs were mutated (Fig. 7B). The insertion of the mammalian minimal promoter (minP), which is suitable for mammalian cells, into the downstream region of the kJHRE reporter (−477 to +1883), did not enhance JH-dependent induction (Supplementary Fig. 3A online). In Drosophila S2 cells, the kJHRE reporter was significantly activated by JH only when the both TcMet and TcSRC were expressed and this induction was abolished by the use of a mutated kJHRE reporter (Supplementary Fig. 3B online).
Discussion
In the current study, we established a novel cell line (Tc81) from T. castaneum embryos. This highly sought after cell line was highly efficient in RNAi and could be used for the analysis of JH-dependent signaling pathways.
In T. castaneum, RNAi-mediated knockdown of a target gene by direct injection of dsRNA into individuals has been shown to provide systemic and highly efficient knockdown throughout the embryonic and postembryonic stages27. Therefore, we hypothesized that a cell line generated from T. castaneum tissues would also maintain the characteristic of high RNAi efficiency. Although one T. castaneum cell line was established very recently from adult and pupal tissues6, the efficiency of RNAi in these cells has not yet been determined. In this study, we generated a new T. castaneum cell line (Tc81) with a different origin (embryos) and demonstrated that this cell line maintained high RNAi efficiency, as expected. The cells exhibited a rapid decline in Met transcripts in a gene-specific manner following a single soaking with low concentrations of Met dsRNA. Repeated dsRNA treatment was not required to obtain highly efficient knockdown through day 8 after dsRNA treatment. Thus, Tc81 cells appear to be valuable for RNAi-based functional gene analysis.
Tc81 cells were shown to be highly sensitive to JH in terms of the induction of the Kr-h1 gene. The Kr-h1 gene has been reported to be induced by JH in a wide variety of insect species16,19,20,21,26,28,29,30 and also in several insect cell lines25,26. Therefore, Kr-h1 is recognized as an indicator of the JH sensitivity of individual insects and insect cells. In Tc81 cells, TcKr-h1 transcripts were rapidly induced by JHA (within 30 min) and by subnanomolar concentrations of natural JHs. This high JH sensitivity implied that Tc81 cells may be valuable for the analysis of the JH signaling pathway at the cellular level.
Moreover, the dose-response and temporal JH-induction profiles of TcKr-h1 suggested that Tc81 cells maintained some characteristic features of JH responsiveness observed in T. castaneum individuals. In terms of TcKr-h1 induction, Tc81 cells were more sensitive to JH III (EC50, 2.7 × 10−11 M) than to JH I (EC50, 4.7 ×10−11 M). In contrast, NIAS-Bm-aff3 cells derived from B. mori fat bodies are more sensitive to JH I (EC50, 1.2 × 10−10 M) than to JH III (EC50, 2.6 × 10−10 M)26. Since JH I and JH III are the major JHs in Lepidoptera and Coleoptera, respectively31, Tc81 and NIAS-Bm-aff3 cells reflect the characteristic response to JH species in their respective organisms. In addition, the temporal JH-induction profile of the Kr-h1 gene in Tc81 was similar to that observed in T. castaneum pupae19. TcKr-h1 reached its peak at 1 h and decreased soon after in T. castaneum pupa that were topically treated with JHA19. Likewise, TcKr-h1 was induced in 1 h and then declined rapidly by 2 h in Tc81 cells. In contrast, in B. mori NIAS-Bm-aff3 cells, Kr-h1 transcripts reached a peak at 2 h after JHA treatment and were maintained at high levels until 12 h after treatment26. Therefore, the early decline in TcKr-h1 after JH induction observed both in pupae and cells may be regulated by a common mechanism at the cellular level.
Taking advantage of the high RNAi efficiency and JH sensitivity of Tc81 cells, we established the function of TcMet and TcSRC in JH signaling. Previously, using in vivo RNAi in the larvae and pupae of T. castaneum individuals, we found that TcKr-h1 is induced by JH via Met19. However, since systemic RNAi was employed in T. castaneum individuals, it was unclear whether the effects of RNAi were tissue autonomous or occurred via humoral and/or neuronal regulation from other tissues in which the target gene was knocked down. In addition, in vivo RNAi is limited in that it cannot be used for the functional analysis of vital genes whose knockdown results in a lethal phenotype. For example, SRC RNAi in T. castaneum larvae caused a lethal phenotype32, which hampered the elucidation of its function in JH signaling. Thus, the use of RNAi in Tc81 cells could circumvent these disadvantages of in vivo RNAi. Indeed, our study revealed that both TcMet and TcSRC mediated JH signaling to induce TcKr-h1 in a cell-autonomous manner, demonstrating the usefulness of this novel cell line.
In a previous study, we reported the presence of a kJHREc-like sequence upstream of TcKr-h126. In the current study, we identified an additional putative kJHREc in the first intron of TcKr-h1. By reporter assays in Tc81 cells, we demonstrated that both kJHREcs were critical for the JH-dependent induction of the TcKr-h1 gene. Intriguingly, additional kJHREc-like sequences were also found in the first intron of Kr-h1 genes in B. mori, A. mellifera, and D. melanogaster. Recently, Shin et al. reported an E-box-like sequence (CACGCG), which interacts with the protein complex of Met and Cycle, upstream of the Kr-h1 (AaKr-h1) gene in A. aegyptii33. This E-box-like sequence was also found in the JHRE of the early trypsin gene24, but was not found in the promoter or first intron region of the TcKr-h1 gene examined in this study (Supplementary Fig. 4 online). On the other hand, a kJHREc-like sequence (GCCTCCACGTG) was found in the vicinity of the E-box-like sequence in AaKr-h133. Since Kr-h1 is commonly induced by JH in a wide variety of insect species as described above, it is reasonable to hypothesize that the critical role of kJHREc for JH-dependent transcriptional induction is highly conserved in insects.
The presence of multiple Kr-h1 transcript isoforms that differ in their 5′ regions were reported in D. melanogaster, B. mori and Frankliniella occidentalis15,26,30. In D. melanogaster and B. mori, the α-isoform is transcribed from a promoter located upstream of the first exon and the β-isoform is transcribed from an alternative promoter located in the first intron15,26. Therefore, the kJHRE upstream of the first exon and in the first intron may regulate the α- and β-isoforms, respectively, in these insects. Alternatively, kJHREc in the first intron may work together with the upstream kJHREc to induce Kr-h1 transcripts from the upstream promoter. We have attempted to isolate the second isoform of TcKr-h1, for which transcription starts downstream of kJHREc in the first intron, but our attempts using RACE have not yet been successful. Further analyses of the role of individual kJHREcs in the isoform-specific regulation of Kr-h1 are required in T. castaneum and other insect species.
In combination with reporter assays and RNAi in Tc81 cells, we clearly demonstrated that TcMet and TcSRC were involved in the JH-dependent induction of TcKr-h1 via interactions with the kJHRE. We have previously shown that BmMet2 and BmSRC induce BmKr-h1 via interaction with the kJHRE using reporter assays in HEK293 cells heterologously expressing BmMet2 and BmSRC26. However, since RNAi has not been shown to be effective in B. mori individuals and cells, we failed to demonstrate direct interactions between kJHRE, BmMet2 and BmSRC in B. mori cells26. Thus, in the current study, we used a combination of reporter assays and RNAi in Tc81 cells to facilitate the analysis of JH signaling. This strategy in Tc81 cells may be more generally applicable to the analysis of interactions between transcription factors and cis-regulatory elements.
Finally, we attempted to reconstitute the JH induction system using kJHRE, TcMet and TcSRC in mammalian HEK293 cells lacking an intrinsic JH signaling pathway. Using partial recombinant proteins containing PAS domains, previous studies have shown that TcMet and TcSRC physically interact with each other depending on the presence of JH23,25. The 2-hybrid assay system in HEK293 cells using full-length TcMet and TcSRC proteins in the current study was able to confirm the JH-dependent interaction of these proteins, indicating that both proteins were functional in mammalian cells. However, contrary to our expectations, the heterologous expression of full-length TcMet and TcSRC proteins in HEK293 cells could only marginally activate a reporter carrying 2 kJHREc sequences, unlike the case of BmMet2 and BmSRC proteins26. In contrast, in Drosophila S2 cells, JH-dependent induction was observed only in the context of TcMet and TcSRC expression. These results suggested that additional factors intrinsic to insect cells are vital for the JH-dependent activation of the TcKr-h1 gene via kJHREc (Fig. 7C). Using Tc81 cells, RNAi-based screening of the missing factors guided by kJHRE reporter activity is currently underway.
In conclusion, we have succeeded in establishing a new T. castaneum cell line and elucidated a portion of the JH signaling pathway in T. castaneum using this novel cell line. Since reverse genetic analysis using injection of dsRNA in vivo is highly effective in T. castaneum, T. castaneum is frequently used as a model organism to elucidate the molecular mechanisms of various physiological phenomena, such as hormonal regulation, immunity, apoptosis and RNAi7. We anticipate that our novel Tc81 cell line will become a vital tool applicable to a wide range of research fields in T. castaneum.
Methods
Establishment of a T. castaneum cell line (Tc81)
A wild-type strain of T. castaneum, which was obtained from the National Food Research Institute (Japan), was reared in whole wheat flour at 30°C. Twenty eggs were sterilized within 24 h of being laid by immersion in 1% Kitchen Haiter solution (Kao) for 30 min and 70% ethanol for 5 min. The sterilized eggs were rinsed twice with Carlson's solution34 and once with MGM-464 medium supplemented with 20% fetal bovine serum (FBS)35. The rinsed eggs were gently homogenized with a pestle by hand and the homogenized cells were cultured at 25°C with gentle shaking in 600 μL MGM-464 medium containing 20% FBS, 1 mg/mL reduced-form glutathione and 1% antibiotic-antimycotic (Gibco, Invitrogen) for 5 days. Fresh medium (500 μL) was then added to the culture and the shaking culture was changed to a static culture. The medium was partially replaced (30%) with fresh medium once a week. After the fifth generation of culture, the medium was changed to MGM-464 medium containing 10% FBS (glutathione- and antibiotic-free) and this process of adding and replacing medium was continued for 15 months. Unless stated otherwise, MGM-464 medium supplemented with 10% FBS was used for the following experiments. The average sizes of vesicles and cells were calculated by measuring the lengths of 18 vesicles in Fig. 1 and 5 cells in the inset in Fig. 1, respectively. The nuclei were stained with VECTASHIELD Mounting Medium with DAPI (Vector Laboratories). For dsRNA treatment and reporter assays, healthy vesicles were collected by centrifugation at 300 × g for 3 min and pellets were disrupted by gentle pipetting 5 times. To obtain growth curves for Tc81 cells, the cells were seeded at the 120th passage at a density of 1.27 × 108 cells/well in 100 μL medium in 96-well plates (Iwaki) and were collected every other day for 8 days. The collected cells were centrifuged at 500 × g for 3 min and were lysed with cell lysis buffer from the ProteoJET Cytoplasmic and Nuclear Protein Extraction Kit (Fermentas). The lysed cells were stained with VECTASHIELD Mounting Medium with DAPI (Vector Laboratories) and DAPI-stained nuclei were counted under a fluorescence microscope; the number of nuclei was assumed to represent the number of cells.
Chemicals
JH I was purchased from SciTech, JH III was purchased from Sigma-Aldrich and FA was purchased from Echelon Research Laboratories. Methoprene (SDS Biotech) was a kind gift from Dr. Syo Sakurai (Kanazawa University). Appropriate amounts of compounds dissolved in methanol were transferred into glass vials coated with polyethylene glycol 20,000 (PEG20,000; Wako). The methanol was evaporated under a stream of nitrogen, appropriate amounts of medium were added to the vial and the compounds were dissolved by sonication using a Biorupter (Cosmo Bio).
JH treatment in Tc81 cells
To examine temporal changes in the expression of TcKr-h1 after methoprene (JHA) treatment, Tc81 cells were seeded at a density of 1.0 × 103 vesicles with 100 μL medium in a well of 96-well plate and incubated for 1 day before JHA treatment. Fresh medium (100 μL) containing 20 μM JHA was added to precultured cells (final concentration of JHA, 10 μM) and the cells were cultured for 30 min to 24 h at 25°C and collected for RNA extraction.
To examine the concentration-response relationship, cells were seeded at 1.0 × 103 vesicles/well in 100 μL medium in 96-well plates coated with PEG20,000 and incubated for 1 day before JH treatment. Fresh medium containing JH I, JH III, JHA, or FA was added to precultured cells and the cells were incubated for 2 h at 25°C and collected for RNA extraction.
cDNA cloning of TcSRC
Primers used for the cloning of TcSRC cDNA were designed based on the sequence of the T. castaneum genome (Supplementary Table 1 online). A TcSRC cDNA fragment was amplified by PCR with first-strand cDNA prepared from the whole bodies of T. castaneum larvae and RTPCR_FW and RTPCR_RV primers (Supplementary Table 1 online). The 5′- and 3′-end sequences were obtained by 5′ RACE and 3′ RACE with 5′ RACE and 3′ RACE primers, respectively (Supplementary Table 1 online) using a GeneRacer kit (Invitrogen). The full-length cDNA sequence was obtained by combining the sequence data obtained from RT-PCR and RACE analyses. The full ORF of TcSRC was amplified from the cDNAs by PCR using ORF_F and ORF_R primers (Supplementary Table 1 online), subcloned into the pCR4Blunt-TOPO vector (Invitrogen) and sequenced.
RNAi experiments
Template DNA fragments of TcMet, TcSRC and TcKr-h1 used for the synthesis of double-stranded RNA (dsRNA) were amplified by PCR with primers and templates listed in Supplementary Table 2 online and purified using a Wizard SV Gel and PCR Clean-Up System (Promega). dsRNAs were synthesized from the amplified DNA using RiboMAX SP6 and T7 Large Scale RNA Production Systems according to the manufacturer's instructions (Promega).
To examine the temporal effect of RNAi by soaking cells in dsRNAs, Tc81 cells were seeded at a density of 1.0 × 103 vesicles in 100 μL medium in a well of a 96-well plate and dsRNA for MalE or TcMet (1 μL, 5 μg/μL) was added to the medium (final concentration, 50 ng/μL). The cells were cultured for 1–8 days after the first dsRNA treatment. In some experiments, dsRNA for MalE or TcMet (1 μL, 5 μg/μL) was further added to the cells at day 4 after the first dsRNA treatment (final concentration, 100 ng/μL) and incubated for an additional 1–4 days.
To examine concentration-response relationships, cells were seeded at a density of 1.0 × 103 vesicles/well in 100 μL medium containing various concentrations of dsRNAs for MalE or TcMet (0.0005–500 ng/μL) in 96-well plates. Cells were then incubated at 25°C for 60 h and collected for RNA extraction.
Quantitative RT-PCR (qPCR)
qPCR analysis was performed essentially as described previously26. The primers used for qPCR are listed in Supplementary Table 2 online.
Identification of the kJHREc in Kr-h1 genes of several insect species
We searched for putative kJHRE core sequences (kJHREcs) by homology using the sequence 5′-CTCCACGTG or its complementary sequence 5′-CACGTGGAG as queries in the first intron of Kr-h1 genes of 4 insect species (T. castaneum, B. mori, Apis mellifera and D. melanogaster).
Construction of reporter and expression vectors
Genomic DNA was extracted from T. castaneum larvae by conventional methods36. A region containing the 5′-flanking and first intron (−477 to +1833) of the TcKr-h1 gene (Supplementary Fig. 4 online) was amplified from genomic DNA by PCR using the KOD FX DNA polymerase (Toyobo, Japan) and gene-specific primers tagged with attB1 or attB2 (listed in Supplementary Table 3 online). The amplified DNA was inserted into the pGL4.14 luciferase reporter vector (Promega) modified for the Gateway system (Invitrogen)26 to construct pGL4.14_−477/+1883. Deletion and mutation vectors were constructed from pGL4.14_−477/+1883 by inverse PCR (iPCR) using the KOD -Plus- Mutagenesis Kit (Toyobo) with TcKrh1_ProiPCR primers (Supplementary Table 3 online).
Vectors for expressing TcMet or TcSRC proteins fused at the N terminus with the GAL4 DNA-binding domain (GAL4DBD) or VP16 activation domain (VP16AD) were constructed using the CheckMate Mammalian Two-Hybrid System (Promega). Full ORFs of TcMet and TcSRC cDNAs were amplified by PCR using the templates and primers listed in Supplementary Table 1 online. The amplified DNAs were ligated into NotI- and KpnI-digested pBIND and pACT plasmids (Promega). To create vectors that expressed native TcMet and TcSRC proteins, GAL4DBD was removed from pBIND_TcMet and pBIND_TcSRC plasmids by iPCR with the pBINDiPCR primers listed in Supplementary Table 1 online.
Transfection and reporter assays
Tc81 cells were seeded at a density of 1.0 × 103 vesicles/well in 96-well plates and transfected with 40 μL medium (IPL-41, Gibco, Invitrogen) containing a mixture of plasmid DNAs (a reporter vector and a reference vector carrying Renilla luciferase, pIZT_RLuc37, 0.2 μg each) and a transfection reagent (0.6 μL; Fugene HD, Promega). Fresh medium (100 μL MGM-464 medium containing 10% FBS) was added to the wells 1 h after the transfection. In reporter assays with RNAi, dsRNA (500 ng), plasmid DNAs (0.2 μg each) and transfection reagent (0.6 μL) were mixed with 40 μL IPL-41 medium and used for transfection. HEK293 cells were seeded at a density of 0.2 × 105 cells/well in 96-well plates and cultured for 2 days before transfection. HEK293 cells were then transfected with 40 μL medium (MEM, Sigma-Aldrich) containing a mixture of plasmid DNAs (a reporter vector and a reference vector carrying Renilla luciferase, pRL-TK [Promega], 0.2 μg each) and Lipofectamine 2000 (0.6 μL; Invitrogen). The medium was replaced with MEM supplemented with 10% FBS and nonessential amino acids (Gibco, Invitrogen) 6 h after transfection. The transfected cells were then incubated for 60 h and treated with JHs for 1 day. Reporter activities were measured using a Dual-Luciferase Reporter Assay System (Promega) and a luminometer (ARVO; PerkinElmer) according to the manufacturers' instructions.
References
Zettler, J. L. & Cuperus, G. W. Pesticide resistance in Tribolium castaneum (Coleoptera, Tenebrionidae) and Rhyzopertha dominica (Coleoptera, Bostrichidae) in wheat. J. Econ. Entomol. 83, 1677–1681 (1990).
Phillips, T. W., Jiang, X. L., Burkholder, W. E., Phillips, J. K. & Tran, H. Q. Behavioral responses to food volatiles by 2 species of stored-product Coleoptera, Sitophilus oryzae (Curculionidae) and Tribolium castaneum (Tenebrionidae). J. Chem. Ecol. 19, 723–734 (1993).
Young, M., Beeman, R. W. & Arakane, Y. RNAi-based functional genomics in Tribolium castaneum and possible application for controlling insect pests. Entomol. Res. 42, 1–10 (2012).
Richards, S. et al. The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955 (2008).
Lorenzen, M. D. et al. piggyBac-based insertional mutagenesis in Tribolium castaneum using donor/helper hybrids. Insect Mol. Biol. 16, 265–275 (2007).
Goodman, C. L. et al. A cell line derived from the red flour beetle Tribolium castaneum (Coleoptera: Tenebrionidae). In Vitro Cell. Dev. Biol. Anim. 48, 426–433 (2012).
Smagghe, G., Goodman, C. L. & Stanley, D. Insect cell culture and applications to research and pest management. In Vitro Cell. Dev. Biol. Anim. 45, 93–105 (2009).
Riddiford, L. M. Cellular and molecular actions of juvenile hormone .1. General considerations and premetamorphic actions. Adv. Insect Physiol. 24, 213–274 (1994).
Gilbert, L. I., Granger, N. A. & Roe, R. M. The juvenile hormones: historical facts and speculations on future research directions. Insect Biochem.Mol. Biol. 30, 617–644 (2000).
Riddiford, L. M. Juvenile hormone: the status of its "status quo" action. Arch. Insect Biochem. Physiol. 32, 271–286 (1996).
Riddiford, L. M. Juvenile hormone action: a 2007 perspective. J. Insect Physiol. 54, 895–901 (2008).
Jindra, M., Palli, S. R. & Riddiford, L. M. The juvenile hormone signaling pathway in insect development. Annu. Rev. Entomol. 58, 181–204 (2012).
Wilson, T. G. & Fabian, J. A Drosophila melanogaster mutant resistant to a chemical analog of juvenile hormone. Dev. Biol. 118, 190–201 (1986).
Ashok, M., Turner, C. & Wilson, T. G. Insect juvenile hormone resistance gene homology with the bHLH-PAS family of transcriptional regulators. Proc. Natl. Acad. Sci. U. S. A. 95, 2761–2766 (1998).
Pecasse, F., Beck, Y., Ruiz, C. & Richards, G. Krüppel-homolog, a stage-specific modulator of the prepupal ecdysone response, is essential for Drosophila metamorphosis. Dev. Biol. 221, 53–67 (2000).
Minakuchi, C., Zhou, X. & Riddiford, L. M. Krüppel homolog 1 (Kr-h1) mediates juvenile hormone action during metamorphosis of Drosophila melanogaster. Mech. Dev. 125, 91–105 (2008).
Konopova, B. & Jindra, M. Juvenile hormone resistance gene Methoprene-tolerant controls entry into metamorphosis in the beetle Tribolium castaneum. Proc. Natl. Acad. Sci. U. S. A. 104, 10488–10493 (2007).
Parthasarathy, R., Tan, A. J. & Palli, S. R. bHLH-PAS family transcription factor methoprene-tolerant plays a key role in JH action in preventing the premature development of adult structures during larval-pupal metamorphosis. Mech. Dev. 125, 601–616 (2008).
Minakuchi, C., Namiki, T. & Shinoda, T. Krüppel homolog 1 an early juvenile hormone-response gene downstream of Methoprene-tolerant, mediates its anti-metamorphic action in the red flour beetle Tribolium castaneum. Dev. Biol. 325, 341–350 (2009).
Lozano, J. & Belles, X. Conserved repressive function of Krüppel homolog 1 on insect metamorphosis in hemimetabolous and holometabolous species. Sci Rep 1, 163 (2011).
Konopova, B., Smykal, V. & Jindra, M. Common and distinct roles of juvenile hormone signaling genes in metamorphosis of holometabolous and hemimetabolous insects. PLoS One 6, e28728 (2011).
Miura, K., Oda, M., Makita, S. & Chinzei, Y. Characterization of the Drosophila Methoprene-tolerant gene product. Juvenile hormone binding and ligand-dependent gene regulation. FEBS J. 272, 1169–1178 (2005).
Charles, J. P. et al. Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc. Natl. Acad. Sci. U. S. A. 108, 21128–21133 (2011).
Li, M., Mead, E. A. & Zhu, J. Heterodimer of two proteins mediates juvenile hormone-induced gene expression. Proc. Natl. Acad. Sci. U. S. A. 108, 638–643 (2011).
Zhang, Z., Xu, J., Sheng, Z., Sui, Y. & Palli, S. R. Steroid receptor co-activator is required for juvenile hormone signal transduction through a bHLH-PAS transcription factor, methoprene tolerant. J. Biol. Chem. 286, 8437–8447 (2011).
Kayukawa, T. et al. Transcriptional regulation of juvenile hormone-mediated induction of Krüppel homolog 1, a repressor of insect metamorphosis. Proc. Natl. Acad. Sci. U. S. A. 109, 11729–11734 (2012).
Tomoyasu, Y. & Denell, R. E. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev. Genes Evol. 214, 575–578 (2004).
Zhu, J. S., Busche, J. M. & Zhang, X. Identification of juvenile hormone target genes in the adult female mosquitoes. Insect Biochem. Mol. Biol. 40, 23–29 (2010).
Shpigler, H. et al. The transcription factor Krüppel homolog 1 is linked to hormone mediated social organization in bees. BMC Evol. Biol. 10, 120 (2010).
Minakuchi, C., Tanaka, M., Miura, K. & Tanaka, T. Developmental profile and hormonal regulation of the transcription factors broad and Krüppel homolog 1 in hemimetabolous thrips. Insect Biochem. Mol. Biol. 41, 125–134 (2011).
Goodman, W. G. & Cusson, M. The juvenile hormones. in Insect Endocrinology (ed Gilbert, L. I.). Ch. 8, 310–363 (Academic PressLondon 2012).
Bitra, K., Tan, A., Dowling, A. & Palli, S. R. Functional characterization of PAS and HES family bHLH transcription factors during the metamorphosis of the red flour beetle, Tribolium castaneum. Gene 448, 74–87 (2009).
Shin, S. W., Zou, Z., Saha, T. T. & Raikhel, A. S. bHLH-PAS heterodimer of methoprene-tolerant and Cycle mediates circadian expression of juvenile hormone-induced mosquito genes. Proc. Natl. Acad. Sci. U. S. A. 109, 16576–16581 (2012).
Mitsuhashi, J. Compositions of salt solutions and culture media. in Invertebrate Tissue Culture Methods (ed Mitsuhashi, J.). 401–420 (Springer Lab Manual, Tokyo 2002).
Mitsuhashi, J. Development of highly nutritive culture media. In Vitro Cell. Dev. Biol. Anim. 37, 330–337 (2001).
Sambrook, J., Fritsch, E. F. & Maniatis, T. in Molecular Cloning. 2, 9.14–19 (Cold Spring Harbor Laboratory Press, New York 1989
Kanamori, Y. et al. A eukaryotic (insect) tricistronic mRNA encodes three proteins selected by context-dependent scanning. J. Biol. Chem. 285, 36933–36944 (2010).
Acknowledgements
We thank Dr. Chieka Minakuchi for critical reading of the manuscript. This work was supported by the NIAS Strategic Research Fund (T.K. and T.S.).
Author information
Authors and Affiliations
Contributions
T.K., K.T. and T.S. designed the research, T.K. and K.T. performed the experiments, T.K., K.T. and T.S. analyzed data and T.K. and T.S. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary info
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Kayukawa, T., Tateishi, K. & Shinoda, T. Establishment of a versatile cell line for juvenile hormone signaling analysis in Tribolium castaneum. Sci Rep 3, 1570 (2013). https://doi.org/10.1038/srep01570
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01570
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.