Abstract
Herein we developed a new “smart” Gd-based MR contrast agent (i.e., 1) which is susceptive to furin, a protease overexpressed in tumor. Under the action of furin, 1 condenses to form dimers (1-Ds) and the latter self-assemble into gadolinium nanparticles (Gd-NPs). Relaxivity of 1-D is more than 2 folds of those of 1 and magnevist at 1.5 T and 1.4 folds of that of 1 at 3 T. Intracellular condensation of 1 in furin-overexpressed MDA-MB-468 cells was proven with direct two-photon laser microscopy (TPLM) fluorescence imaging of the cells incubated with the europium analog of 1 (i.e., 2). Intracellular Gd-NPs of 1 were uncovered and characterized for the first time. MRI of MDA-MB-468 tumors showed that 1 has enhanced MR contrast within the tumors than that of its scrambled control 1-Scr.
Similar content being viewed by others
Introduction
Molecular imaging (MI) is a growing research discipline aims at developing and testing novel tools, reagents and methods to detect unique in vivo “biochemical signatures” that differentiate and characterize tissues beyond and before their gross anatomical features becoming obvious1,2. To date, imaging modalities of MI that most commonly used for extracting molecular information are nuclear, MRI and optical techniques3. Among them, MRI has become increasingly popular in experimental MI and clinical radiology because it allows the interrogation of intact, opaque organisms in three dimensions at cellular resolution (~10 μm)4. About 35% of all clinical MR scans utilize contrast agents (CAs). In proton MR, gadolinium (Gd3+)-based T1 CAs are used for reducing the spin-lattice relaxation times of nearby water, increasing the signal from these protons and making the effected voxel seem “brighter” in T1-weighted image. Superparamagnetic iron oxide nanoparticle-based T2 CAs are used to reduce the spin-spin relaxation time of water, make a “negative” contrast effect in T2*/T2-weighted image5. In general, T1-weighted sequences provide images of higher resolution and signal-to-noise ratio than T2*/T2-weighted ones and are free of image artifacts. However, due to the low sensitivity of the CAs, high concentration (0.1–0.6 mM) of CA is always required for a typical MR scanning and this calls for the design of highly potent molecular CAs for success6,7. Common strategy for increasing the longitudinal molar relaxivity (r1) of T1 CA is to prolong its rotational correlation time (τr i.e., the tumbling time of the CA in the water bulk). To achieve this goal, Gd-based agents with higher molecular weights such as Gd functionalized polymer, peptide amphiphiles or viral caspid, dendrimer, liposomes, nanoparticles, micelles, zeolites, fullerenes, carbon nanotubes, clays and quantum dots were prepared and explored8,9,10,11,12. Nevertheless, these pre-made gadolinium complexes are facing the problem of cell membrane translocation and targeting, besides the difficulty and reproducibility of their fabrications13. Therefore, design of “smart” or “activable” MR CAs that modulate their MR properties (e.g., relaxivities) on site upon molecular target interaction will overcome the shortcomings of MRI from bottom up. Unfortunately, up to date, only a few gadolinium-based smart MR probes have been developed, including those responsive to β-galactosidase or myeloperoxidase4,14,15,16,17. Self-assembly, a prevalent and important process in nature18, provides an easy approach to design “smart” MR probes. In brief, it is not difficult for a small molecular probe (i.e., building block for self-assembly) to overcome the barrier of cell membrane and be delivered to the targeting site inside cell. At the targeting site, the building blocks (i.e., small molecular probes) “smartly” start to self-assemble into nano/micro structures with higher molecular weights which are crucial for a T1 CA. Recently, Rao and co-workers developed a biocompatible condensation reaction between 1,2-aminothiol group of cysteine and the cyano group of 2-cyanobenzothiazole (CBT) which could be controlled by pH, reduction and protease at such low a concentration as micromolar for self-assembling nanoparticles with diameters ranging from 8 nm to 170 nm in vitro and in cells19. Using this system, Rao and co-workers have successfully developed the smart MR CAs of first generation which are susceptive to reducing agents (e.g., glutathione in cells) and have enhanced T1 relaxivity more than 100%20. Inspired by this, herein we designed the second generation of smart molecular MRI CAs which are not only responsive to intracellular glutathione (GSH) but also cleavable by intracellular protease furin who is overexpressed in cancer cells. Using this smart CA, we successfully achieved enhanced MRI of MDA-MB-468 tumors on nude mice under common clinical field strength (3 Tesla).
The trans-Golgi protease furin is a kind of protein convertase that plays important roles in homeostasis and in diseases ranging from Alzheimer’s disease to anthrax and Ebola fever and cancer21. Several cancers upregulate furin, including non-small-cell lung carcinomas, squamous-cell carcinomas of the head and neck and glioblastomas22. Moreover, the increase of furin in tumors correlates with an increase of membrane type 1-matrix metalloproteinase (MT1-MMP), one of furin’s substrates. MT1-MMP activates extracellular pro-MMP2 to induce rapid tumor growth and metastasis23. Thus, overexpression of furin offers people with a useful hint of early development of certain cancers. There is one big advantage for chemists to study furin that it preferentially cleaves Arg-X-Lys/Arg-Arg↓X motifs, where Arg is arginine, Lys is lysine, X can be any amino acid residue and ↓ indicates the cleavage site24.
Inspired by these, as shown in Fig. 1, we designed Acetyl-Arg-Val-Arg-Arg-Cys(StBu)-Lys(Gd-DOTA)-CBT (1) for self-assembling gadolinium nanoparticles (Gd-NPs) under the action of furin in living tumor cells. In brief, 1 contains a RVRR peptide sequence for furin cleavage and cell membrane translocation, disulfided Cys for supplying the 1,2-aminothiol group for condensation, Lys conjugated with Gd-DOTA for MRI. After entering cells, the disulfide bond of the Cys motif of 1 is reduced by the intracellular GSH and subsequently its RVRR motif is cleaved by furin on the site of this enzyme (i.e., Golgi body), resulting in the active intermediate 1-Core. Two 1-Cores condense quickly to yield the amphiphilic dimer (i.e., 1-D) which has a hydrophobic macrocyclic core for self-assembling Gd-NPs via π-π stacking among each others. As-formed Gd-NPs should greatly increase the local concentration of Gd inside cells on one hand. On the other hand, the higher molecular weight of the 1-D should have relaxivity enhancement compared with that of 1 at an identical Gd concentration, presumably owing to an increased rotational correlation time. In this work, we demonstrated the furin-controlled condensation of 1 and self-assembly of Gd-NPs in vitro, measured the enhanced relaxivities of the condensation product of 1 (i.e., 1-D), directly visualized the intracellular condensation of 2 with TPLM cell imaging and characterized the Gd-NPs of 1 inside cells for the first time. We also successfully imaged MDA-MB-468 tumors on nude mice with this second generation of smart molecular CA (i.e., 1).
Shematic illustration of a furin-controlled condensation and self-assembly of Gd-NPs in cancer cells.
After entering cancer cells, the disulfide bond of probe 1 is reduced by GSH and the RVRR peptide sequence is cleaved by furin to yield the active intermediate 1-Core. Two 1-Cores condense to yield amphiphilic dimer 1-D which self-assembles into Gd-NPs at or near the locations of furin in cells (i.e., Golgi bodies).
Results
Synthesis
We began the study with the synthesis and preparation of five compounds: 1 and 1-Scr, 2 and 2-Scr and 1-D (Fig. 2 & Supplementary information). The synthesis for these five compounds is simple and straightforward (Supplementary information). Briefly, the Ac-Arg-Val-Arg-Arg-Cys(StBu)-Lys-OH (A) peptide sequence with protection groups was synthesized with Solid Phase Peptide Synthesis (SPPS), then coupled with CBT, purified with high performance liquid chromatography (HPLC) to yield B. Deprotection of B yields C after HPLC purification. Coupling of C with DOTA(OtBu)3 yields D and subsequent deprotection of D with trifluoroacetic acid (TFA) yields E. At pH value of 6–7, 10 equiv. of GdCl3·6H2O chelates with E at room temperature (RT) for 3 h yields 1 after HPLC purification. Synthesis of 1-Scr is similar to that of 1 with A used for the synthesis of 1 being replaced by peptide sequence Ac-Arg-Lys-Arg-Cys(StBu)-Arg-Val-OH (F). Syntheses of 2 and 2-Scr are similar to those of 1 and 1-Scr with GdCl3·6H2O used at the last steps for the syntheses of 1 or 1-Scr being replaced with EuCl3·6H2O respectively. Following the literature, we synthesized NH2-Cys(SEt)-Lys(Gd-DOTA)-CBT (K)20. Reduction of K with 4 equiv. of tris(2-carboxyethyl)phosphine (TCEP) yields 1-D after HPLC purification.
Chemical structures of the five designed probes.
1, 1-Scr and 1-D are Gd-based T1 MR CAs. 1 is susceptive to furin, while 1-D is the condensation product of 1 after furin cleavage. 1-Scr is the scrambled control probe of 1. 2 is the Eu analog of 1 for TPLM cell imaging. 2-Scr is the Eu analog of 1-Scr.
Furin-controlled condensation of 1 and self-assembly of Gd-NPs and nanocharacterizations
To test our hypothesis, we used 1 for in vitro study. As shown in Fig. 3a, after 17 h incubation of 1 at 100 μM and 30°C with 1 nmol/U of furin, we directly injected the incubation mixture into a HPLC system and collected the peaks for matrix-assisted laser desorption/ionization (MALDI) mass spectroscopic analysis. Interestingly, peaks on HPLC traces at retention times of 38.2 min (1-D-1, 19.8%), 39.5 min (1-D-2, 21.2%), 40.5 min (1-D-2, 20.0%) and 42.8 min (1-D-4, 13.5%) share an identical molecular weight and were identified as the condensation products of 1 (i.e., 1-D, Supplementary Fig. S1). Owing to the presence of L-lysine, these four peaks probably represent the four diastereoisomers of 1-D that arise from two different ring-closing orientations during the condensation between L-cysteine motif and cyano group of 125. These four dimers account for 74.5% of the enzymatic products of 1 in total. UV-Vis spectrum at 500–700 nm of the above reaction mixture showed an obvious increase of absorption compared with that of the solution without furin, suggesting the formation of nanostrcutures (Supplementary Fig. S2). Directly taking the above dispersion for scanning electron microscope (SEM) and transmission electron microscope (TEM) observation, we uncovered the 3-dimensional and 2-dimensional depositions of the Gd-NPs of 1 (Fig. 3b&c). The Gd-NPs have uniform spherical shapes and an average diameter of 57.1 ± 11.9 nm.
Characterizations of furin-controlled condensation and self-assembly of Gd-NPs of 1 in vitro.
(a) Upper, HPLC trace of 1 in water; lower, HPLC trace of the incubation mixture of 1 at 100 μM after incubation with 1 nmol/U of furin at 30°C for 17 h. (b) SEM and (c) TEM images of the Gd-NPs of 1 in the above incubation mixture.
Measurement of longitudinal molar relaxivity (r1)
The MR contrast properties of 1, 1-Scr and 1-D were evaluated in vitro together with commercial CA Gd-DTPA (Magnevist) as control using phosphate buffered phantoms (pH 7.4, 0.2 M). T1-weighted MR phantom imaging of these CAs was conducted on both 1.5 Tesla (T) and 3 T MRI scanners (Supplementary Fig. S3–12). Plots of signal intensity versus inversion time give the T1 relaxation times of each CAs at certain concentrations. We calculated the longitudinal molar relaxivity (r1) of each CA according to the equation r1 = ΔR1/[CA], where the relaxation rate R1 is 1/T1. As shown in Fig. 4a, at 1.5 T and RT, the r1s were determined to be 6.00 s−1mM−1 for 1, 7.42 s−1mM−1 for 1-Scr and 13.24 s−1mM−1 for 1-D respectively. Relaxivities of Magnevist at this condition were determined to be 5.39 s−1mM−1, which agree well with those reported in literature26. The relaxivity of 1-D at 1.5 T (13.24 s−1mM−1) is 2-fold more than that of 1 and comparable to those protein-bound Gd-DOTA analogues reported by Caravan et al27. Measurement of the relaxivities of abovementioned MR CAs at 3 T is shown in Fig. 4b. Similar to those at 1.5 T, 1-D has the highest relaxivity and that of Magnevist is the lowest. In general, relaxivities of these MR CAs at 3 T are lower than those at 1.5 T. All the data of their relaxivities at 1.5 T or 3 T are summarized in Table 1.
T1 relaxivity measurements of 1, 1-Scr and 1-D.
Spin-lattice 1/T1 relaxation rates of 1, 1-Scr and 1-D at different concentrations in phosphate buffer (pH 7.4, 0.2 M) at 1.5 T (a) and 3 T (b), compared to the commercially available MR CA (Magnevist).Relaxivity rates r1 were obtained by comparing the measured (symbols) and theoretical (lines) values.
Direct imaging furin-controlled intracellular condensation of 2 with TPLM
As the Eu-analog of 1, 2 has all the necessaries for furin-controlled intracellular condensation. Luminescence of europium enables this intracellular process of 2 visible under a TPLM. Before applying 2 or 2-Scr for TPLM cellular imaging, we tested the expression level of furin in human breast cancer cell line MDA-MB-468 with western blot and immunofluorescence staining. Human colon carcinoma LoVo cells were chosen as control cell lines for western blot study because they are reported to be furin-deficient28. The protein cell lysates prepared from these two cell lines were analyzed with western blot. Using glycolytic enzyme glyceraldehydes phosphate dehydrogenase (GAPDH) as control, as shown in Fig. 5a, MDA-MB-468 cells revealed a positive signal for furin while very weak signal of furin was detected in LoVo cells. Quantification of the western blot signals with image J (NIH, USA) indicated that furin in MDA-MB-468 cell has an expression level of 96.2% of GAPDH while that in LoVo is 24.5% of GAPDH (4-fold, p = 0.008) (Fig. 5b). High expression of furin in MDA-MB-468 cells was also confirmed with immunofluorescence staining of furin using rhodamine-labelled secondary antibody (Fig. 5c). An overlay of the fluorescence staining of furin with that of nucleus staining (4',6-diamidino-2-phenylindole, DAPI staining) clearly shows that the locations of furin (i.e., the Golgi bodies) as reported (Fig. 5d)21. TPLM imaging of MDA-MB-468 cells incubated with 2 at 100 μM for 8 h shows strong fluorescence signals similar to those in Fig. 5c (Fig. 5e & Supplementary Movie S1), suggesting 2 was under the action of furin and trapped at/near the locations of furin. Interestingly, TPLM imaging of MDA-MB-468 cells incubated with 2-Scr at same condition only exhibits uniform, weak fluorescence signals (Fig. 5f).
Expression of furin in MDA-MB-468 cells and two-photon laser microscopy images of MDA-MB-468 cells incubated with 2 or 2-Scr.
Western blot analysis (a) and quantification (b) of furin in MDA-MB-468 cells and LoVo cells. Furin was highly expressed in MDA-MB-468 cells (96.2% of GAPDH) while in LoVo cells it was less expressed (24.5% of GAPDH, 4-fold, p = 0.008). (c, d) Immunofluorescence staining of MDA-MB-468 cells with rhodamine-labeled antibody against furin: DsRed channel (red, furin) (c); merged image with DAPI (blue, nucleus) (d). Scale bar: 20 μm. (e, f) TPLM images (λex = 725 nm, λem = 565–636 nm) of MDA-MB-468 cells incubated with 2 (e) or 2-scr (f) at 100 μM for 8 h and then rinsed and fixed prior to imaging. Scale bar: 20 μm.
Self-assembly of Gd-NPs of 1 in MDA-MB-468 cells
After validation of the intracellular condensation of 2 upon furin cleavage in MDA-MB-468 cells, we used electron microscope (EM) to localize the Gd-NPs self-assembled from the condensation products of 1 incubated with the cells. Cells treated with 1-Scr were studied in parallel because 1-Scr is inactive to furin. Before EM observation, furin protein expression in the cells was quantified with western blot after 8 h incubation of the cells with the probes. Unexpectedly, as shown in Fig. 6a, after 8 h incubation with 1 at 100 μM, furin protein expression in MDA-MB-468 cells exhibits an obvious decrease compared with that in the cells untreated. Quantification of the western blot signals indicated that furin protein in cells treated with 1 has an expression level of 41.7% of GAPDH while that in cells untreated is 93.9% of GAPDH (2.3-fold, p = 0.001) (Fig. 6b). In contrast, cells treated with 1-Scr did not show decreased expression level of furin protein, compared with that in cells untreated (Supplementary Fig. S13). After 8 h incubation with 1 or 1-Scr, the cells were fixed with 2.5% glutaraldehyde and thin sections of cells were cut and mounted on copper grids for EM observation. For MDA-MB-468 cells that treated with 1, large area of clustered Gd-NPs at/near the sites of Golgi bodies were clearly observed (Fig. 6c). High magnification of the EM image indicated that these intracellular Gd-NPs of 1 have an average diameter of 24.0 ± 2.3 nm (Fig. 6d), much smaller than those formed in vitro (Fig. 3b). In contrast, there are no Gd-NPs presented in MDA-MB-468 cells treated with or without 1-Scr (Supplementary Fig. S14&15).
Expression of furin in MDA-MB-468 cells before and after incubation with 1 and electron microscopy images of the cells after 8 h incubation with 1.
Western blot analysis (a) and quantification (b) of furin expression levels in MDA-MB-468 cells before and after incubation with 1 at 100 μM for 8 h. Expression of furin in cells treated with 1 has an obvious decrease (41.7% of GAPDH), compared with that in cells untreated (93.9% of GAPDH, 2.3-fold, p = 0.001). Low (c) and high (d) magnification Electron microscopic images of MDA-MB-468 cells after incubation with 1 at 100 μM for 8 h. Large area of clustered Gd-NPs of 1 were found at/near Golgi bodies. Scale bar in c: 2 μm. Scale bar in d: 400 nm.
MRI of MDA-MB-468 tumors with 1
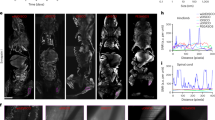
Having shown that 1 selectively condenses and self-assembles into Gd-NPs in furin-overexpressed MDA-MB-468 cells, coronal MR images of mice with subcutaneous MDA-MB-468 cell xenografts were acquired precontrast and at various times after the first intravenous (i.v.) injections of 1 (1st injection: 0.15 mmol/kg at 0 min; 2nd injection: 0.15 mmol/kg at 50 min) (Fig. 7a&b and Supplementary Fig. S16). In the precontrast images (i.e., 0 min), there was little intrinsic contrast between the implanted MDA-MB-468 tumors and surrounding muscle. At 50 min following the administration of the 1st dose of 1, significantly increased enhancement was observed within the MDA-MB-468 tumors (49.1% increase of grey value compared to that at 0 min). By 90 min (i.e., 40 min after the 2nd dose of injection), slightly enhanced signal to that at 50 min could be observed (53.9% increase of grey value compared to that at 0 min) and by 240 min the signal within the tumors remained obvious (20.5% increase of grey value compared to that at 0 min, Supplementary Fig. S16). To assess specificity of 1, control experiments were performed by two i.v. injections of 1-Scr into mice with subcutaneous MDA-MB-468 cell xenografts at the exactly same doses and time points to those of 1. In these animals, enhancement in contrast signal within tumors was also observed. However, at each of the time points studied, the signal was clearly lower than that of mice injected with 1 (Fig. 7a). Quantitative analysis of the MR images is presented in Fig. 7b. These results indicated that 1 is obviously better than 1-Scr as a MR CA for imaging MDA-MB-468 tumors, probably that 1 is susceptive to furin while 1-Scr is not. Since furin protein levels were decreased in MDA-MB-468 cells in vitro treated with 1 (Fig. 6a&b), we also quantified furin protein expression in MDA-MB-468 tumors in vivo with western blot after MRI. As shown in Fig. 7c, after 240 min of MRI, expression level of furin in MDA-MB-468 tumors treated with 1 exhibited an obvious decrease compared with that in control groups. In contrast, expression of furin in tumors treated with 1-Scr did not show obvious change compared with that in control groups. Quantification of the western blot signals indicated that furin in tumors of mice injected with 1 has an expression level of 54.8% of GAPDH while that in tumors of mice untreated is 90.9% of GAPDH (1.7-fold, p = 0.002) (Fig. 7d). Furin in tumors of mice injected with 1-Scr has an expression level of 106.3% of GAPDH, no obvious difference from that in tumors of mice untreated (p = 0.08). To directly observe furin expression levels, after 240 min of MRI, the tumors were excised for immunofluorescence staining and imaging. The results are shown in Fig. 7e. Unlike those of mice injected with or without 1-Scr which show connective red fluorescence signal (i.e., staining of furin) among the nucleuses (DAPI staining), tumor sections of mice injected with 1 exhibited disconnected, attenuated red signal, which also indicates the down regulation of furin protein level by the probe. Although the furin protein itself has lower expression in mice injected with 1, ICP-MS analysis indicated that at 240 min after the first injection of 1, tumors on mice have an average Gd content of 0.12 μg/g, 2.6-fold high of that of tumors treated with 1-Scr (0.046 μg/g) (Supplementary Table S1), suggesting that 1 is a very potent probe for imaging MDA-MB-468 tumors in vivo. Other organs (lung, liver, spleen and kidney) except brain in mice treated with 1, all exhibit higher contents of Gd than those treated with 1-Scr (Supplementary Table S1). Hematoxylin and eosin (HE) staining of the tissue slices of mice treated with 1 or 1-Scr did not show pathologic changes compared with those of mice untreated, suggesting that the doses of 1 or 1-Scr herein injected for MRI did not result in toxicity to the mice within the time window of imaging (Supplementary Fig. S17).
In vivo imaging MDA-MB-468 tumors with 1.
(a) Representative coronal MR images of mice with subcutaneously xenografted MDA-MB-468 tumors at 0 min, 50 min and 90 min after two intravenous injections of 1 (upper) or 1-Scr (lower) via tail veins (1st injection: 0.15 mmol/kg at 0 min; 2nd injection: 0.15 mmol/kg at 50 min). Tumors are indicated by arrows. (b) Quantitative analysis of grey values of tumor MR images at various time. (c) Western blot analysis and (d) quantification of furin expression in MDA-MB-468 tumors on mice injected with 1, with/without 1-Scr after 240 min of MRI. Furin protein in tumors of mice injected with 1 has a decreased expression level of 54.8% of GAPDH, compared with that in tumors on mice untreated (90.9% of GAPDH, 1.7-fold, p = 0.002). Expression of furin protein in tumors of mice injected with 1-Scr does not show obvious change (106.3% of GAPDH), compared with that in tumors on mice untreated (p = 0.08). (e) Immunofluorescence staining images of tumors on mice untreated (Control), injected with 1 or 1-Scr. Furin is stained red and nucleuses are stained blue with DAPI. Scale bar: 20 μm.
Discussion
1-Scr is an isomer of 1 but with a scrambled peptide sequence which could not be cleaved by furin. Compound 2 is the europium (Eu) analog of 1. We designed 2 to evaluate the cell permeability and demonstrate the intracellular condensation of 1 by directly imaging the intracellular behavior of 2 with TPLM because 2 has luminescence emissions at 594 nm and 616 nm when excited with two-photon excitation at 725 nm (Supplementary Fig. S18). In parallel, we synthesized 2-Scr to mimic the intracellular behavior of 1-Scr with TPLM imaging. Since the majorities of the condensation products of 1 upon furin cleavage are 1-Ds, we also synthesized 1-D for in vitro study.
In vitro relaxivity measurement results indicated that both 1 and 1-Scr have relaxivities higher than that of Magnevist. This probably dues to that the molecular weight of 1 or 1-Scr (1645 Da) is higher than that of Magnevist (662 Da). Interestingly, although 1-D has a molecular weight (1860 Da) close to that of 1, its r1 is 2-fold more higher than that of 1, near 2-fold of that of 1-Scr and near 3-fold of that of Magnevist. This might be ascribed to that the hydrophobic macrocyclic ring of 1-D increases its rotational correlation time (τr) in aqueous solution, suggesting that 1 is a “smart” MR contrast agent susceptive to furin.
Since 2 and 2-Scr are the europium analogs of 1 and 1-Scr respectively, their intracellular behavior should represent that of 1 or 1-Scr accordingly. For 2-Scr, it is the diastereoisomer of 2. Therefore, it should have similar cell permeability to that of 2, proven by the TPLM imaging of 2 and 2-Scr on furin-deficient LoVo cells (Supplementary Fig. S19). The difference between the two TPLM images (i.e., Fig. 5e&f) suggests that 2 could be intracellularly cleaved by furin and condense at the sites of furin while 2-Scr could not be trapped inside the cells because 2-Scr is not susceptible to furin. To our best of knowledge, caspases and cathepsins are also able to hydrolyze a peptide substrate to yield a N-terminal cysteine motif. However, they have their own specific substrates for cleavage instead of RVRR (e.g., DEVD for caspase-3 and ZVKM for cathepsin B)29,30. Therefore, the specificity of 1 (or 2) to furin for condensation should be much higher than those to other proteases. Interestingly, the intracellular Gd-NPs observed inside the cells after incubation with 1 are much smaller than those obtained via in vitro incubation of 1 with furin (24.0 nm vs. 57.1 nm). We suspected that this might due to high viscosity of cytosol and the intracellular fibrous networks of the cells hindering the small Gd-NPs from further aggregation.
Preliminary results of in vivo imaging MDA-MB-468 tumors with 1 were obtained. MRI results indicate that 1 is obviously better than 1-Scr for MDA-MB-468 tumor imaging even in the mice with lower expression of furin protein, which further suggests that 1 is a powerful probe. The big difference of Gd contents in the organs (lung, liver, spleen and kidney) except brain between the mice treated with 1 and 1-Scr at 240 min (Supplementary Table S1) might be ascribed to the structural difference between 1 and 1-Scr. Detailedly, cleavage of the amide bond between the Cys and Arg motifs of 1 by the proteinase (furin or other proteinases) in these organs results in condensation reaction and formation of the Gd-NPs which trap the agent in the tissues/organs. In contrast, even the amide bond between the Cys and Arg motifs of 1-Scr is cleaved by the proteinase resulting in the condensation and formation of nanoparticles, the Gd-DOTA motif of 1-Scr is excluded from neither the condensation reaction nor the nanoparticle formation thereafter. Thus, it is conceivable that the Gd-DOTA motif cleaved from 1-Scr should have a smaller molecular weight and be excreted from the body within a very short time window (the plasma half-life for Gd-DOTA is 90 min in patient with normal renal function). HE staining results indicate that the MR CAs designed in our work are biocompatible, suggesting that 1 could be likely developed for clinical trial in the near future.
In summary, taking advantage of a biocompatible condensation, we have successfully developed the second generation of new smart Gd-based MR contrast agent (i.e., 1) for imaging in vitro and in vivo. 1 is susceptive to furin, a protease overexpressed in tumor. Upon furin cleavage, 1 condenses to form amphiphilic dimer (1-D) and the latter self-assembles into Gd-NPs thereafter. Relaxivity of 1-D is more than 2-fold of that of 1. Using the europium analog of 1 (i.e., 2), we directly imaged the intracellular condensation of 2 in furin-overexpressed MDA-MB-468 cells. By incubating the cells with 1 for 8 h, for the first time to our best of knowledge, we uncovered and characterized the intracellular Gd-NPs self-assembled from the condensation products of this small molecule uptaken by the cells. Compared with that of its scrambled control probe 1-Scr, 1 showed enhanced MR contrast within MDA-MB-468 tumors. Immunofluorescence staining of the tumors indicated that it is furin to trap 1 in tumors. Encouraged by these exciting results above, we envisioned that more “smart” probes based on other proteases overexpressed in tumors could also be invented for tumor imaging, using this versatile condensation platform or other “click chemistry” techniques31.
Methods
General methods
All the starting materials were obtained from Adamas or Sangon Biotech. Commercially available reagents were used without further purification, unless noted otherwise. All other chemicals were reagent grade or better. Furin was purchased from Biolabs (2,000 UmL−1); one unit (U) is defined as the amount of furin that releases 1 pmol of methylcoumarinamide (MCA) from the fluorogenic peptide BOC-RVRR-AMC (Bachem) in one minute at 30°C. 1H NMR spectra were obtained on a 300 MHz Bruker AV 300. MALDI-TOF/TOF mass spectra were obtained on a time-of-flight Ultrflex II mass spectrometer (Bruker Daltonics), HPLC analyses were performed on an Agilent 1200 HPLC system equipped with a G1322A pump and in-line diode array UV detector using a YMC-Pack ODS-AM column with CH3OH (0.1% of TFA) and water (0.1% of TFA) as the eluent. SEM images were obtained on JEOL-JSM-6700F electron microscope at an accelerating voltage of 5.0 KV. TEM images were obtained on a JEOL 2010 electron microscope, operating at 100 KV. The cryo-dried samples were prepared as following: a copper grid coated with carbon was dipped into the suspension solvent and placed into a vial, which was plunged into liquid nitrogen until no bubbles were apparent. Then water was removed from the frozen specimen by a freeze-drier. ICP-AES measurements were conducted on an ICP-96B machine equipped with a PGS-2 atomic emission spectrometer (Zeiss). ICP-MS measurements were conducted on an X Series 2 machine (Thermo Fisher Scientific).
Cell culture
MDA-MB-468 human breast adenocarcinoma epithelial cells and Human colon carcinoma LoVo cells were cultured in Dulbecco’s modified eagle medium (GIBCO) supplemented with 10% fetal bovine serum (FBS, GIBCO).
Two photon laser microscopy
MDA-MB-468 or LoVo cells were cultured on the glass slide, incubated with 2 or 2-scr at 100 μM for 8 h, washed with phosphate buffered saline (PBS) for three times, fixed with 4% paraformaldehyde at RT for 30 min, washed with PBS a further three times and once with distilled water. Then the cells were mounted with 50% glycerol and imaged under a Zeiss 710 confocal laser-scanning microscope equipped with a Coherent Mrux1 titanium: sapphire mode-locked laser. The excitation wavelength for TPLM was 725 nm (2 × 362.5 nm = 725 nm). A 565–636 nm band pass filter was used for cell imaging.
Electron microscopic imaging
MDA-MB-468 cells were incubated with 1 or 1-scr at 100 μM for 8 h, washed for three times with phosphate-buffered saline (PBS), fixed with 2.5% glutaraldehyde at RT for 30 min. The cells were then detached from culture dishes, centrifuged (300 rpm, 15 min) and washed with PBS for a further three times and then stained with 1% OsO4 in double-distilled water for 1.5 h. Then the cells were dehydrated in ethanol and embedded in Epon. Thin sections (80 nm) were cut and mounted on copper grids, stained with saturated solution of uranyl acetate and lead citrate for electron microscope observation.
In vitro and in vivo MRI
The in vitro phantom MR experiments were performed on a 1.5 T (Simens, Magnetom-essenza) and 3 T (Simens, Trio-Tim) scanners, using a head RF coil. The scanning procedure began with a localizer and then consisted of a series of inversion-prepared fast spin echo images, identical in all aspects (TR 1740 ms, TE 13, BW 140 kHz, percent phase field of view 50, slice thickness 3 mm, matrix 136 × 136, NEX 1) except for the inversion time (TI) which was varied as follows: 1500, 1200, 1000, 800, 500, 400, 200, 150, 100 and 75 ms. Signal intensity (SI) versus TI relationships were fit to the following exponential T1 decay model by non-linear least squares regression: SI(TI) = A1*exp(−TI/T1) + SI(0). Relaxation rates (R1) were determined as 1/T1. Longitudinal molar relaxivities (r1, units of s−1mM−1) were calculated as the slope of R1 vs [CA] after the determination of true Gd concentration of each sample by the ICP-AES or ICP-MS measurement. The in vivo MR imaging of MDA-MB-468 tumor xenografted nude mice was performed on 3 T scanner (Simens, Trio Tim), using head RF coil. Female mice of 3–4 weeks old were provided by Sun Yat-sen University Laboratory Animal Center (Guangzhou, China). MDA-MB-468 tumor lesions were established by subcutaneous dorsal flank injection of 4×107 tumor cells in 100 μL PBS for each mouse. Visible tumors were normally observed 2–4 weeks after injection. The tumor-xenografted mice were then subjected randomly into two groups for tail vein injections of 1 or 1-scr (1st injection: 0.15 mmol/kg at 0 min; 2nd injection: 0.15 mmol/kg at 50 min). The images were taken at a time sequence from 0 min to 240 min using T1-weighted MR acquisition sequence with the following parameters: TR 2000 ms, TE 70, BW 289 kHz, percent phase field of view 60, slice thickness 2 mm, matrix 144 × 144, NEX 1. The intensity of MR signal in tumor for each test was determined by standard region-of-interest measurement with Image J.
Western blot
MDA-MB-468 cells were incubated with 1 or 1-Scr at 100 μM for 8 h, then washed with ice-cold PBS for three times and harvested in 1.5 mL eppendorf tubes respectively, followed by centrifugation at 2,000 g and 4°C for 10 min. The supernatants were removed and the cell samples were treated with radioimmunoprecipitation assay (RIPA) lysis buffer containing 4% protease inhibitor (Roche). Cells in the mixture were broken by sonifier cell disruptor (200 W, 6 s) and lysed for 3 min on ice. Cell extracts were clarified by centrifugation at 12,000 g and 4°C for 15 min and then mixed with SDS sample buffer for denaturation at 100°C for 10 min. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to Immun-Blot polyvinylidene fluoride (PVDF) membrane (Bio-Rad). Western blotting was carried out using anti-furin (1:1200, Sigma Aldrich) or GAPDH (1:2000, Cell Signaling Technology) at 4°C overnight and horse radish peroxidase (HRP)-conjugated secondary antibodies at room temperature for 1 h. All antibodies were used in 5% skim milk (BD Bioscience). All experiments were carried out at least in triplicate. For in vivo assay, mice were sacrificed after MR scanning and the tumors were removed and treated with RIPA lysis buffer containing 4% protease inhibitor (Roche). The tissue mixtures were broken by sonifier cell disruptor (400 W, 10 s) and lysed for 3 min on ice. Extracts were clarified by centrifugation at 12,000 g and 4°C for 15 min and then mixed with SDS sample buffer for denaturation at 100°C for 10 min. The protocol of western blot analysis was described above.
ICP-AES measurement
After MR scanning, phantom samples were diluted with water until the calculated concentrations of Gd3+ were within the range of 1–20 ppm. The exact concentration of Gd3+ in each phantom sample was determined with a standard calibration curve using standard Gd3+ samples at concentrations of 1, 5, 10 and 20 ppm.
ICP-MS measurement
After 240 min of T1-weighted MRI, tumor-bearing nude mice were sacrificed. Tissues and organs including lungs, brains, livers, spleens, kidneys and tumors of these mice were collected and weighted. After that, each of the tissues was soaked in 5.0 mL of nitric acid (70%), heated for at least 6 h until the liquid was totally evaporated and the tissue was completely digested. The residue was dissolved in 2% nitric acid until the concentration of Gd3+ was within 1–100 ppb. Exact concentration of Gd3+ in each tissue sample was determined by comparing with the standard Gd3+ samples at 50 ppb.
References
Weissleder, R. & Mahmood, U. Molecular imaging. Radiology 219, 316–333 (2001).
Winter, P. M. et al. Molecular Imaging of angiogenesis in nascent vx-2 rabbit tumors using a novel alpha(v)beta(3)-targeted nanoparticle and 1.5 tesla magnetic resonance imaging. Cancer Res. 63, 5838–5843 (2003).
Baker, M. The whole picture. Nature 463, 977–980 (2010).
Louie, A. Y. et al. In vivo visualization of gene expression using magnetic resonance imaging. Nat. Biotechnol. 18, 321–325 (2000).
Chou, S. W. et al. In Vitro and in Vivo Studies of FePt Nanoparticles for Dual Modal CT/MRI Molecular Imaging. J. Am. Chem. Soc. 132, 13270–13278 (2010).
Major, J. L. & Meade, T. J. Bioresponsive, Cell-Penetrating and Multimeric MR Contrast Agents. Acc. Chem. Res. 42, 893–903 (2009).
Terreno, E., Castelli, D. D., Viale, A. & Aime, S. Challenges for Molecular Magnetic Resonance Imaging. Chem. Rev. 110, 3019–3042 (2010).
Bridot, J. L. et al. Hybrid gadolinium oxide nanoparticles: Multimodal contrast agents for in vivo imaging. J. Am. Chem. Soc. 129, 5076–5084 (2007).
Karfeld-Sulzer, L. S. et al. Protein Polymer MRI Contrast Agents: Longitudinal Analysis of Biomaterials In Vivo. Magn. Reson. Med. 65, 220–228 (2011).
Chen, W. T. et al. Dynamic Contrast-Enhanced Folate-Receptor-Targeted MR Imaging Using a Gd-loaded PEG-Dendrimer-Folate Conjugate in a Mouse Xenograft Tumor Model. Mol. Imaging Biol. 12, 145–154 (2010).
Yang, H. et al. Detection of a Family of Gadolinium-Containing Endohedral Fullerenes and the Isolation and Crystallographic Characterization of One Member as a Metal-Carbide Encapsulated inside a Large Fullerene Cage. J. Am. Chem. Soc. 130, 17296–17300 (2008).
Ananta, J. S. et al. Geometrical confinement of gadolinium-based contrast agents in nanoporous particles enhances T-1 contrast. Nat. Nanotechnol. 5, 815–821 (2010).
Silva, G. A. Neuroscience nanotechnology: Progress, opportunities and challenges. Nat. Rev. Neurosci. 7, 65–74 (2006).
Bogdanov, A., Jr, Matuszewski, L., Bremer, C., Petrovsky, A. & Weissleder, R. Oligomerization of paramagnetic substrates result in signal amplification and can be used for MR imaging of molecular targets. Mol. imaging 1, 16–23 (2002).
Querol, M. & Bogdanov, A. Amplification strategies in MR imaging: Activation and accumulation of sensing contrast agents (SCAs). J. Magn. Reson. Imaging 24, 971–982 (2006).
Chen, J. W., Pham, W., Weissleder, R. & Bogdanov, A. Human myeloperoxidase: A potential target for molecular MR imaging in atherosclerosis. Magn. Reson. Med. 52, 1021–1028 (2004).
Ronald, J. A. et al. Enzyme-Sensitive Magnetic Resonance Imaging Targeting Myeloperoxidase Identifies Active Inflammation in Experimental Rabbit Atherosclerotic Plaques. Circulation 120, 592–599 (2009).
Whitesides, G. M., Mathias, J. P. & Seto, C. T. Molecular Self-Assembly and Nanochemistry - a Chemical Strategy for the Synthesis of Nanostructures. Science 254, 1312–1319 (1991).
Liang, G. L., Ren, H. J. & Rao, J. H. A biocompatible condensation reaction for controlled assembly of nanostructures in living cells. Nat. Chem. 2, 54–60 (2010).
Liang, G. et al. Controlled Self-Assembling of Gadolinium Nanoparticles as Smart Molecular Magnetic Resonance Imaging Contrast Agents. Angew. Chem. Int. Ed. 50, 6283–6286 (2011).
Thomas, G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3, 753–766 (2002).
Mbikay, M., Sirois, F., Yao, J., Seidah, N. G. & Chretien, M. Comparative analysis of expression of the proprotein convertases furin, PACE4, PC1 and PC2 in human lung tumours. Br. J. Cancer 75, 1509–1514 (1997).
Sounni, N. E. et al. Expression of membrane type 1 matrix metalloproteinase (MT1-MMP) in A2058 melanoma cells is associated with MMP-2 activation and increased tumor growth and vascularization. Int. J. Cancer 98, 23–28 (2002).
Hosaka, M. et al. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J. Biol. Chem. 266, 12127–12130 (1991).
Ye, D., Liang, G., Ma, M. L. & Rao, J. Controlling Intracellular Macrocyclization for the Imaging of Protease Activity. Angew. Chem. Int. Ed. 50, 2275–2279 (2011).
Stanisz, G. J. & Henkelman, R. M. Gd-DTPA relaxivity depends on macromolecular content. Magn. Reson. Med. 44, 665–667 (2000).
Overoye-Chan, K. et al. EP-2104R: A fibrin-specific gadolinium-based MRI contrast agent for detection of thrombus. J. Am. Chem. Soc. 130, 6025–6039 (2008).
Lehmann, M. et al. Deficient processing and activity of type I insulin-like growth factor receptor in the furin-deficient LoVo-C5 cells. Endocrinology 139, 3763–3771 (1998).
Hook, V., Hook, G. & Kindy, M. Pharmacogenetic features of cathepsin B inhibitors that improve memory deficit and reduce beta-amyloid related to Alzheimer's disease. Biol. Chem. 391, 861–872 (2010).
Cao, C., Chen, Y., Wu, F., Deng, Y. & Liang, G. Caspase-3 controlled assembly of nanoparticles for fluorescence turn on. Chem. Commun. 47, 10320–10322 (2011).
Zeng, Y., Ramya, T. N. C., Dirksen, A., Dawson, P. E. & Paulson, J. C. High-efficiency labeling of sialylated glycoproteins on living cells. Nat. Methods 6, 207–209 (2009).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21175122, 91127036, 81071207 and 81271622), the Fundamental Research Funds for Central Universities (WK2060190018 and 10ykjcll) and Anhui Provincial Natural Science Foundation (1108085J17). The authors are grateful to H.F. Zhang for fruitful discussions and the Center for Integrative Imaging (CII) of Hefei National Laboratory for Physical Science at the Microscale for the imaging facilities.
Author information
Authors and Affiliations
Contributions
Chun-Yan Cao and Ying-Ying Shen contributed equally to this work.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Movie S1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Cao, CY., Shen, YY., Wang, JD. et al. Controlled intracellular self-assembly of gadolinium nanoparticles as smart molecular MR contrast agents. Sci Rep 3, 1024 (2013). https://doi.org/10.1038/srep01024
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01024
This article is cited by
-
Iron(III)-Tannic Molecular Nanoparticles Enhance Autophagy effect and T1 MRI Contrast in Liver Cell Lines
Scientific Reports (2018)
-
Applications of CBT-Cys click reaction: past, present, and future
Science China Chemistry (2018)
-
Deciphering the Biochemical Pathway and Pharmacokinetic Study of Amyloid βeta-42 with Superparamagnetic Iron Oxide Nanoparticles (SPIONs) Using Systems Biology Approach
Molecular Neurobiology (2018)
-
A Microfluidic Platform to design crosslinked Hyaluronic Acid Nanoparticles (cHANPs) for enhanced MRI
Scientific Reports (2016)
-
Molecular imaging of enzyme activity in vivo using activatable probes
Science Bulletin (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.