Abstract
hOGG1 encodes a DNA repair enzyme responsible for the excision of reactive oxygen species (ROS) in damaged DNA. Previous studies have obtained inconsistent results. To validate the association between the hOGG1Ser326Cys polymorphism and lung cancer risk, we performed an updated meta-analysis of 20 studies (8739 cases and 10385 controls) using STATA version 11.1. With this approach, we tested the overall and subgroup association between the SNP and lung cancer susceptibility stratified by ethnicity, control sources, cell histotypes and smoking status. We demonstrated a novel, significant correlation between the hOGG1 Ser326Cys polymorphism and increased lung cancer susceptibility in Caucasians. Our findings indicate a need for larger-scale studies to verify the association of this SNP with lung cancer risk in Caucasians.
Similar content being viewed by others
Introduction
Human 8-oxoguanine DNA glycosylase -1 (hOGG1) is located at chromosome 3p26.2. It encodes an enzyme responsible for removing the most common product of oxidative damage in DNA, namely 8-hydroxyguanine (8-OH-G)1. 8-OH-G can induce G→T or A→C base mismatches during DNA replication, thereby possibly triggering the onset of carcinogenesis2,3.
Codon 326 at position 1245 in exon 7 of hOGG1 holds a single nucleotide polymorphism (SNP) with a C→G variation, thereby the amino acid translation of codon 326 can be changed from serine (Ser) to cysteine (Cys)4,5. Experiments have illustrated that the DNA glycosylase encoded by the Cys326 variant exhibits remarkably lower 8-OH-G excision activity than the wild-type Ser326 allele, because the Cys326 variant enzyme has a lower affinity to lesions of damaged DNA than Ser326 enzyme6. Thus, hOGG1 Ser326Cys polymorphism is speculated to associate with multiple types of cancer due to the compromised cleavage of 8-OH-G7.
Lung cancer is the most commonly diagnosed cancer (1.61million diagnoses, 12.7% of the total cancer diagnoses) and is estimated to be the leading cause of cancer death (1.38 million deaths, 18.2% of the total cancer deaths) worldwide8. Numerous investigations have studied the association between the hOGG1 Ser326Cys polymorphism and this malignancy9. However, the results of these studies have been inconsistent, partly due to genetic or other sources of heterogeneity, including differences in eligibility criteria and analysis approaches, small sample sizes, publication biases and exogenous confounders10. For this reason, meta-analyses with robust statistical power have been frequently performed to validate the association between the hOGG1 Ser326Cys polymorphism and the risk of lung cancer. Li et al11 found no association between the hOGG1 Ser326Cys polymorphism and increased risk of lung cancer susceptibility except in Asians, while Kiyohara et al12 found a significant association between the hOGG1 Ser326Cys polymorphism and the risk of lung cancer in the overall population and in an Asian subgroup. In another study, Guan et al13 uncovered a potential trend of significant linkage between hOGG1 Ser326Cys polymorphism and lung cancer risk in Caucasians. Since these publications, more studies of the hOGG1 Ser326Cys polymorphism in relation to lung cancer susceptibility have been completed. Thus, we conducted an updated meta-analysis by adding the latest data and avoiding sample overlapping with the aim of gaining a more reliable evaluation of the association between hOGG1 Ser326Cys polymorphism and lung cancer susceptibility.
Results
Study characteristics
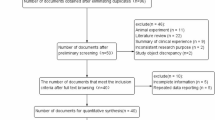
Twenty-eight articles were identified to meet the inclusion criteria. We thoroughly reviewed these articles to detect overlapping samples. Studies by Vogel et al14, Sorensen et al15, Hatt et al16 and Loft et al17,18 were found to share a common sample sources from a Danish prospective follow-up study. Therefore, only the study by Sorensen et al, which had the largest sample size, was used in our meta-analysis. The studies by Sunaga et al19 and Kohno et al20consisted of the same lung adenocarcinoma cases; we included only the study by Kohno et al because it had a larger sample size. The studies by Liang et al21, Zienolddiny et al22 and Liu et al23 were excluded because the genotype distribution among the controls was deviated significantly from HWE (P<0.05). Finally, 20 articles, including 8739 lung cancer cases and 10385 controls were ascertained for use in our meta-analysis15,20,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41.We treated each ethnic population within each paper as a separate study to perform an ethnicity-based subgroup analysis. In a multi-ethnic study by Le et al27, the data were extracted into Asian, Caucasian and Hawaiian subgroups. In the article by Chang et al34, data were separated into Latino and African-American subgroups. Considering the comparability with previous published meta-analyses12,13, we defined data by Karahalil et al33 as Turkish ethnicity in a concordant manner. Eight studies contained population-based controls, while twelve utilised hospital-based controls. Essential characteristics about each original study, HWE values, odd ratio (OR), 95% confidence interval (95%CI) and approaches used to confirm genotyping results are shown in Table 1.
Heterogeneity and model
All heterogeneity statistic I2 values except that in smoking subgroup (I2 = 53.4%) were observed less than 50% in the present study, which indicated that the appropriate pooling model should be fixed effects (Inverse Variance). For the smoking subgroup, a random effects model was used. Furthermore, using a suitable underlying genetic model in genetic association studies is crucial for combining data biologically rather than just statistically. According to the methodology for genetic model selection developed by Thakkinstian et al42, we decided to use the recessive genetic model. After a sensitivity analysis, no individual study was found to affect the overall result robustly, which implied the magnitude of the summary evaluation.
Gene effect
The overall frequency of the Cys allele in the case group was significantly higher than that in the control group (39.7% versus 35.1%, P<0.01). Among the Asian subgroup, the Cys variant frequency was 54.0% in the cases and 51.7% in the controls (P<0.01). More or less, the higher frequency of the Cys allele in cases suggested a potential association of the variant with risk of lung cancer.
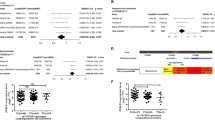
The overall results of the genetic analysis indicated a significant association between the hOGG1 Ser326Cys polymorphism and lung cancer risk (OR = 1.20, 95%CI: 1.10–1.30) (Table 2). In the subgroups by ethnicity, a significant association was observed in Caucasians (OR = 1.32, 95%CI: 1.05–1.67) and in Asians (OR = 1.18, 95% CI: 1.07–1.29) respectively (Figure 1). In the stratified analysis based on control sources, our study also showed a significant linkage of the hOGG1 Ser326Cys polymorphism with lung cancer risk in both population-based (OR = 1.18, 95%CI: 1.04–1.34) and hospital-based controls (OR = 1.21, 95%CI: 1.08–1.35) (Figure 2).
For the study stratified by smoking status, only eight studies were available20,26,29,30,33,6,7,8,9,10,11,12,13,14,15,16,17,18,19,21,22,23,24,25,27,28,31,32,34,35,36,37,38. No correlation was found between hOGG1 Ser326Cys polymorphism and lung cancer among non-smoking (OR = 1.09, 95%CI: 0.92–1.29) or smoking subgroups (OR = 1.24, 95%CI: 0.95–1.61). Another stratified study referring to histological subtypes, due to a lack of well-documented pathological data in most original studies, only ten studies were useful for stratification by the following histological subtypes: small cell carcinoma, squamous cell carcinoma and adenocarcinoma20,24,25,29,30,33,36,37,38,39. Significant association between hOGG1 Ser326Cys polymorphism and an increased risk of lung cancer was found only in lung adenocarcinoma subgroup (OR = 1.31, 95% CI: 1.14–1.51) (Table 3).
The current study also examined the association between the hOGG1 Ser326Cys polymorphism and lung cancer risk, adjusting for both study design and ethnicity. Interestingly, we observed a significant relationship between this SNP and lung cancer susceptibility among smokers in the Asian population (OR = 1.25, 95%CI: 1.04–1.51), but an assessment of Caucasians was not possible due to the limited data available. Similar to the results of the combined analysis, the hOGG1 Ser326Cys polymorphism showed a significant association with lung cancer susceptibility only in the adenocarcinoma cell type subgroups in the Asian and Caucasian populations respectively (Table 4).
The weight of each study contributing to the overall result was calculated. Of the statistical power, 81.83% was from the Asians, while 13.01% was from the Caucasians. The weight of each individual study varied widely, from 22.85% to 0.17%. The hospital-based controls contributed more power than the population-based controls (55.56% versus 44.44%), with the heterogeneity statistic I2 being remarkably higher in the hospital-based control group than that in the population-based controls (41.8% versus 10.4%).
Publication bias
The publication bias was accessed using Begg's (P = 0.303) and Egg's (P = 0.185) tests. The funnel plot displayed a symmetric shape (Figure 3), indicating the absence of a publication bias for both positive and negative or non-significant findings from published studies.
Discussion
Genetic epidemiological studies have proposed that there is a relationship between SNPs and diseases. However, large and well-designed genotype-phenotype investigations with robust statistical power are required to detect these mild to moderate associations. Additionally, there has been increased focus on the modified effects of certain exogenous factors. A predominant DNA glycosylase, encoded by hOGG1, has the ability to recognise and remove an oxidative DNA damage product, namely 8-OH-G. This substance is generally treated as a mutagen because of its ability to induce mutation. Studies have revealed that the Cys-mutant enzyme is less effective at repairing DNA than Ser wild type enzyme6,7,8.
In a published pooled analysis, Li et al reported that there is no relationship between hOGG1 Ser/Cys polymorphism and lung cancer risk11. However, Kiyohara et al found a significant association between this genetic polymorphism and lung cancer by adding several additional case-control studies12. With more studies about hOGG1 and lung cancer were available recently, our updated meta-analysis, which has the largest sample size thus reported, yielded a positive relationship between hOGG1 Ser/Cys polymorphism and lung cancer risk in Caucasians (OR = 1.32, 95%CI: 1.05–1.67). In addition, a recent meta-analysis by Guan et al has predicted a potential connection between the variant and lung cancer in Caucasians13. Our novel finding may be due to an increase in sample sizes and the avoidance of sample overlapping. Another reason may be that we objectively and precisely stratified the population based on ethnicity subgroups. Because the detection for mild to modest risk genetic risk effects requires sufficient statistical power, we proposed that large case-control studies may help to further validate the true association between the genetic variant and lung cancer among Caucasians.
Based on source of controls, we were able to observe that the heterogeneity statistic I2 in the hospital-based subgroup was higher than that in the population-based subgroup (41.8%versus10.4%). To some extent, hospital-based controls were recruited as lung cancer-free individuals regardless of their status concerning other diseases, which might be a potential source of heterogeneity because of a mixture of other diseases, particularly if the disease had effect upon genotyped results11.We suggested that the use of population-based controls should be more representative.
In another analysis, stratified according to smoking status, no significant association was observed between the hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Contrary to this finding, Cys allele has been reported to be associated with lung cancer risk among heavy smokers39. Another study by Li et al identified a marginally increased risk of hOGG1 Ser326Cys polymorphism in non-smoking subjects harbouring the Cys allele11. To the best of our best knowledge, multiple tobacco-related chemicals are capable of inducing DNA mutations and initiating carcinogenesis, especially in lung cancer43. Thus, the complexities of exogenous modification to gene and gene-environment interactions remain a field to be explored. We attributed the inconsistency of different studies to a lack of universal standards when delineating subgroups, including different category criteria such as smoking duration and intensity. Otherwise, the data were primarily obtained from self-reported data from questionnaires of participants, so inaccurate confounders should be taken into account. In addition, the current study employed analyses adjusted for study design and ethnicity and yielded a positive association between the hOGG1condon326 polymorphism and increased lung cancer risk in smoking group among Asians. We assumed that modification of the study design and ethnicity could lower the heterogeneity and reduce the potential of confounders. It also implied the important role of the modified analyses in pooled studies.
Given the diversity of histopathologic categories in lung cancer, stratified evaluation was conducted for small cell carcinoma, squamous cell carcinoma and adenocarcinoma. We found a significant association between the hOGG1 Ser/Cys polymorphism and lung adenocacinoma. Our result was consistent with one former meta-analysis focused on histological types of lung cancer35. Future independent studies are proposed to collect well-documented characteristics of participants incorporating smoking details and histological details by well-trained investigators. The control populations, matched for age, gender and alcohol use, should be twice the size of the case populations in future study.
Our meta-analysis had some limitations. First, a great proportion of statistical power was contributed by the Asian ethnicity, although the subgroup analysis was able to significantly reduce the between-ethnicity heterogeneity. Indeed, more relevant studies of Caucasians are essential. Second, due to the limitation of eligible data, the subgroups based on smoking status were crudely classified into smoking and non-smoking subgroups, regardless of the smoking duration and consumption. Thus, potential sources of heterogeneity were included when the data were combined. Although the interaction between smoking status and hOGG1 condon326 is of great interest, the limited data available for use in the current meta-analysis were not sufficient to identify an association between the genotype and cigarette smoking. Third, the small sample size of lung cancer cell sub-types might have restricted the power of our meta-analysis to reveal a potential connection.
Methods
Eligiblity of relevant studies
All original articles published in English that examined the association of the hOGG1 Ser326Cys polymorphism with lung cancer (published before November 2011) were considered for our meta-analysis. The PubMed, Embase, HuGENet and Ovid databases were searched to identify appropriate studies. The following combinations of terms were used in our database searches: (“Lung cancer” or “Lung Neoplasms” or “Pulmonary Cancer” or “Pulmonary Neoplasms”) and (“polymorphism” or “SNP” or “allele” or “variant”) and (“OGG1” or “hOGG1” or “OGG1 enzyme” or “hOGG1: Human 8-oxoguanine DNA glycosylase-1”). Furthermore, the searches were supplemented by references cited in other papers. Inclusion criteria were: (1) studies assessed linkage of hOGG1 Ser326Cys polymorphism with lung cancer risk; (2) lung cancer cases should be diagnosed explicitly; (3) controls should be unrelated cancer-free individuals. When multiple reports had overlapping sample populations, only the study with largest sample size was retained.
Data extraction
For each available study, the following information was extracted: the first author, year of publication, ethnicity of participants, source of controls, number of genotyped cases/controls, method for quality control of genotyping result, smoking status and histological sub-type. The data were primarily extracted from tables and supplemented by significant information presented in texts and/or figures. Two investigators (Y-L H and D-N Z) handled the data simultaneously and separately.
Statistical analysis
Hardy-Weinberg equilibrium (HWE) was assessed for each study using the chi-square test. Studies were considered to deviate from HWE at P<0.0544. The inconsistency index, I2, was calculated to evaluate the variation among studies owing to heterogeneity (0%–25% was considered to have no heterogeneity; 25%–50% was considered to have moderate heterogeneity; 50%–75% was considered to have large heterogeneity; 75%–100% was considered to have extreme heterogeneity)45. The data were combined using logistic regression with the fixed-effects pooling model if there was no or moderate heterogeneity (I2<50%). Alternatively, the random effects model was used (I2>50%). Sensitivity analysis was performed by excluding one study at a time to determine the corresponding magnitude of the weight of each study to the summary results. The most biologically fit genetic model was selected according to the comprehensive effect of the gene using logistic regression42,46. To extensively explore the genetic heterogeneity, stratified analyses were conducted by ethnicity, source of controls, smoking status and histological sub-types. The association between hOGG1 Ser326Cys polymorphism and lung cancer risk was evaluated using the odds ratio (OR) and the 95% confidence interval (CI). Funnel plots, used to observe the publication bias, were complemented with Egger's regression47and Begg's rank correlation test (P > 0.10)48. The statistical analyses were performed using STATA version 11.1 (Stata Corporation, USA). All p values were two-sided.
References
Loft, S. & Poulsen, H. E. Markers of oxidative damage to DNA: antioxidants and molecular damage. Methods Enzymol 300, 166–184 (1999).
Cheng, K. C., Cahill, D. S., Kasai, H., Nishimura, S. & Loeb, L. A. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A----C substitutions. J Biol Chem 267, 166–172 (1992).
Hoeijmakers, J. H. Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374 (2001).
Boiteux, S. & Radicella, J. P. The human OGG1 gene: structure, functions and its implication in the process of carcinogenesis. Arch Biochem Biophys 377, 1–8 (2000).
Shinmura, K. & Yokota, J. The OGG1 gene encodes a repair enzyme for oxidatively damaged DNA and is involved in human carcinogenesis. Antioxid Redox Signal 3, 597–609 (2001).
Hill, J. W. & Evans, M. K. Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic Acids Res 34, 1620–1632 (2006).
Goode, E. L., Ulrich, C. M. & Potter, J. D. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev 11, 1513–1530 (2002).
Ferlay, J. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 127, 2893–2917 (2010).
Weiss, J. M., Goode, E. L., Ladiges, W. C. & Ulrich, C. M. Polymorphic variation in hOGG1 and risk of cancer: a review of the functional and epidemiologic literature. Mol Carcinog 42, 127–141 (2005).
Hung, R. J., Hall, J., Brennan, P. & Boffetta, P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol 162, 925–942 (2005).
Li, H., Hao, X., Zhang, W., Wei, Q. & Chen, K. The hOGG1 Ser326Cys polymorphism and lung cancer risk: a meta-analysis. Cancer Epidemiol Biomarkers Prev 17, 1739–1745 (2008).
Kiyohara, C., Takayama, K. & Nakanishi, Y. Lung cancer risk and genetic polymorphisms in DNA repair pathways: a meta-analysis. J Nucleic Acids 2010, 701760 (2010).
Guan, P., Huang, D., Yin, Z. & Zhou, B. Association of the hOGG1 Ser326Cys polymorphism with increased lung cancer susceptibility in Asians: a meta-analysis of 18 studies including 7592 cases and 8129 controls. Asian Pac J Cancer Prev 12, 1067–1072 (2011).
Vogel, U. et al. No association between base excision repair gene polymorphisms and risk of lung cancer. Biochem Genet 42, 453–460 (2004).
Sorensen, M. et al. Interactions between the OGG1 Ser326Cys polymorphism and intake of fruit and vegetables in relation to lung cancer. Free Radic Res 40, 885–891(2006).
Hatt, L. et al. OGG1 expression and OGG1 Ser326Cys polymorphism and risk of lung cancer in a prospective study. Mutat Res 639, 45–54 (2008).
Loft, S. et al. Prospective study of 8-oxo-7,8-dihydro-2′-deoxyguanosine excretion and the risk of lung cancer. Carcinogenesis 27, 1245–1250 (2006).
Loft, S. et al. Association between 8-oxo-7,8-dihydroguanine excretion and risk of lung cancer in a prospective study. Free Radic Biol Med 52, 167–172 (2012).
Sunaga, N. et al. Contribution of the NQO1 and GSTT1 polymorphisms to lung adenocarcinoma susceptibility. Cancer Epidemiol Biomarkers Prev 11, 730–738 (2002).
Kohno, T. et al. Association of the OGG1-Ser326Cys polymorphism with lung adenocarcinoma risk. Cancer Sci 97, 724–728 (2006).
Liang, G., Pu, Y. & Yin, L. Rapid detection of single nucleotide polymorphisms related with lung cancer susceptibility of Chinese population. Cancer Lett 223, 265–274 (2005).
Zienolddiny, S. et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis 27, 560–567 (2006).
Liu, C. J. et al. The joint effect of hOGG1 single nucleotide polymorphism and smoking habit on lung cancer in Taiwan. Anticancer Res 30, 4141–4145 (2010).
Sugimura, H. et al. hOGG1 Ser326Cys polymorphism and lung cancer susceptibility. Cancer Epidemiol Biomarkers Prev 8, 669–674 (1999).
Wikman, H. et al. hOGG1 polymorphism and loss of heterozygosity (LOH): significance for lung cancer susceptibility in a caucasian population. Int J Cancer 88, 932–937 (2000).
Ito, H. et al. A limited association of OGG1 Ser326Cys polymorphism for adenocarcinoma of the lung. J Epidemiol 12, 258–265 (2002).
Le Marchand, L., Donlon, T., Lum-Jones, A., Seifried, A. & Wilkens, L. R. Association of the hOGG1 Ser326Cys polymorphism with lung cancer risk. Cancer Epidemiol Biomarkers Prev 11, 409–412 (2002).
Lan, Q. et al. Oxidative damage-related genes AKR1C3 and OGG1 modulate risks for lung cancer due to exposure to PAH-rich coal combustion emissions. Carcinogenesis 25, 2177–2181 (2004).
Park, J., Chen, L., Tockman, M. S., Elahi, A. & Lazarus, P. The human 8-oxoguanine DNA N-glycosylase 1 (hOGG1) DNA repair enzyme and its association with lung cancer risk. Pharmacogenetics 14, 103–109 (2004).
Hung, R. J. et al. Large-scale investigation of base excision repair genetic polymorphisms and lung cancer risk in a multicenter study. J Natl Cancer Inst 97, 567–576 (2005).
Matullo, G. et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis 27, 997–1007 (2006).
De Ruyck, K. et al. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res 631, 101–110 (2007).
Karahalil, B. et al. The association of OGG1 Ser326Cys polymorphism and urinary 8-OHdG levels with lung cancer susceptibility: a hospital-based case-control study in Turkey. Arh Hig Rada Toksikol 59, 241–250 (2008).
Chang, J. S. et al. Base excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African-Americans. Carcinogenesis 30, 78–87 (2009).
Okasaka, T. et al. hOGG1 Ser326Cys polymorphism and risk of lung cancer by histological type. J Hum Genet 54, 739–745 (2009).
Chang, C. H. et al. Interactive effect of cigarette smoking with human 8-oxoguanine DNA N-glycosylase 1 (hOGG1) polymorphisms on the risk of lung cancer: a case-control study in Taiwan. Am J Epidemiol 170, 695–702 (2009).
Miyaishi, A. et al. MUTYH Gln324His gene polymorphism and genetic susceptibility for lung cancer in a Japanese population. J Exp Clin Cancer Res 28, 10 (2009).
Li, Z. et al. Genetic polymorphism of DNA base-excision repair genes (APE1, OGG1 and XRCC1) and their correlation with risk of lung cancer in a Chinese population. Arch Med Res 42, 226–234 (2011).
Kohno, T. et al. Contribution of the TP53, OGG1, CHRNA3 and HLA-DQA1 genes to the risk for lung squamous cell carcinoma. J Thorac Oncol 6, 813–817 (2011).
Janik, J. et al. 8-Oxoguanine incision activity is impaired in lung tissues of NSCLC patients with the polymorphism of OGG1 and XRCC1 genes. Mutat Res 709–710, 21–31(2011).
Qian, B. et al. Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer 73, 138–146 (2011).
Thakkinstian, A., McElduff, P., D'Este, C., Duffy, D. & Attia, J. A method for meta-analysis of molecular association studies. Stat Med 24, 1291–1306 (2005).
Hecht, S. S. Approaches to chemoprevention of lung cancer based on carcinogens in tobacco smoke. Environ Health Perspect 105 Suppl 4, 955–963 (1997).
Guo, S. W. & Thompson, E. A. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48, 361–372 (1992).
Higgins, J. P., Thompson, S. G., Deeks, J. J. & Altman, D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003).
Altman, D. G. & Bland, J. M. Comparing several groups using analysis of variance. BMJ 312, 1472–1473 (1996).
Egger, M., Davey Smith, G., Schneider, M. & Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997).
Begg, C. B. & Mazumdar, M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994).
Acknowledgements
We are thankful to Dr. Zhang Wen (Peking Union Medical College) for language revision and for critical discussion of our study. This study was supported by grants from Key Laboratory Youth Science Funds of Guangxi Research Center Open Program (No. 903011335).
Author information
Authors and Affiliations
Contributions
HYL and ZDN have contributed to the conception and design of the study, the analysis and interpretation of data and the revision of the article. ZDN and WJZ searched and selected the studies, carried out the statistical analysis and drafted and revised the article. LGJ and LJX participated in the statistical analysis and helped to search the trials.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Zhong, D., Li, G., Long, J. et al. The hOGG1Ser326Cys Polymorphism and Increased Lung Cancer Susceptibility in Caucasians: An Updated Meta-Analysis. Sci Rep 2, 548 (2012). https://doi.org/10.1038/srep00548
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00548
This article is cited by
-
Synthesis of homogeneous CaMoO4 microspheres with nanopits for high-capacity anode material in Li-ion battery
Applied Physics A (2018)
-
Polymorphisms of human 8-oxoguanine DNA glycosylase 1 and 8-hydroxydeoxyguanosine increase susceptibility to arsenic methylation capacity-related urothelial carcinoma
Archives of Toxicology (2016)
-
DNA Base-Excision Repair Genes OGG1 and NTH1 in Brazilian Lung Cancer Patients
Molecular Diagnosis & Therapy (2015)
-
Smoking and hOGG1 Ser326Cys polymorphism contribute to lung cancer risk: evidence from a meta-analysis
Tumor Biology (2014)
-
Polymorphisms of excision repair gene XPD Lys751Gln and hOGG1 Ser326Cys might not be associated with hepatocellular carcinoma risk: a meta-analysis
Tumor Biology (2013)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.