Abstract
Nicotine is an important chemical compound in nature that has been regarded as an environmental toxicant causing various preventable diseases. Several bacterial species are adapted to decompose this heterocyclic compound, including Pseudomonas and Arthrobacter. Pseudomonas putida S16 is a bacterium that degrades nicotine through the pyrrolidine pathway, similar to that present in animals. The corresponding late steps of the nicotine degradation pathway in P. putida S16 was first proposed and demonstrated to be from 2,5-dihydroxy-pyridine through the intermediates N-formylmaleamic acid, maleamic acid, maleic acid and fumaric acid. Genomics of strain S16 revealed that genes located in the largest genome island play a major role in nicotine degradation and may originate from other strains, as suggested by the constructed phylogenetic tree and the results of comparative genomic analysis. The deletion of gene hpo showed that this gene is essential for nicotine degradation. This study defines the mechanism of nicotine degradation.
Similar content being viewed by others
Introduction
Significant amounts of nicotine wastes are produced by tobacco factories, resulting in various environmental and health effects1. Microbial organisms play important roles in the tobacco manufacturing process by altering the content of nicotine2,3. Recently, Pseudomonas spp. strains were integrated in the processing of tobacco products and the treatment of tobacco wastes4,5,6,7,8,9,10,11. The Gram-negative soil bacterium P. putida S16 has the ability to degrade nicotine9. The basic steps and corresponding intermediates of nicotine catabolism generating 2,5-dihydroxy-pyridine (DHP) were earlier reported by Wada and Xu6,7,12. A gene cluster (here designated as nic1) containing the genes encoding nicotine oxidoreductase (NicA) and 6-hydroxy-3-succinoylpyridine hydroxylase (HspA) is involved in the catabolism of nicotine to DHP (Fig. 1A) in P. putida S1613,14. A second HSP hydoxylase (HspB), a more active form than HspA with only 11% amino acid sequence homology, was purified and characterized. The hspB gene deletion showed that this gene is essential for nicotine degradation and site-directed mutagenesis identified an FAD binding domain15. However, despite the work described above, our understanding is still incomplete, especially the late steps of nicotine degradation. Recently, the genome sequence of strain S16 was completed16 and it helps us to further identify the genomic and metabolic diversity of this species (Fig. S1). In addition, a DHP oxidase in nicotinic acid degradation from P. putida N-9 was earlier purified and crystallized by Rittenberg and coworkers17. Moreover, Jimenez et al. identified and characterized a gene cluster (nic genes) responsible for the aerobic nicotinic acid degradation in P. putida KT244018. The mechanism of DHP transformation may be similar during nicotine degradation and nicotinic acid degradation in Pseudomonas. All the above information may be helpful in data mining and identification of related genes in the late steps of nicotine degradation.
Chemical reactions and genes involved in the nicotine degradation pathway of P. putida S16.
(A) Pyrrolidine pathway of nicotine degradation. The reactions performed by the enzymes encoded by the nic2 gene cluster are inside the red-dashed box. (B) Genetic organization of the nic2 cluster of P. putida S16 as compared with similar gene clusters from other bacteria. HspB, HSP hydroxylase (black); Iso, maleate isomerase (red); Nfo, NFM deformylase (green); Hpo, DHP dioxygenase (orange); Ami, maleamate amidase (blue); Hna, 6-hydroxynicotinate 3-monooxygenase (dark cyan); Orfx, no predicted function (grey). The percentages represent amino acid homology of related enzymes.
This study reports a gene cluster (here designated as nic2) encoding for 5 factors that mediate in the conversion of HSP to fumarate, of which the hspB gene is included. The late steps of the nicotine degradation pathway in P. putida S16 was first proposed to be from DHP through the intermediates, N-formylmaleamic acid, maleamic acid and maleic acid, to fumaric acid, as indicated in the red dashed box in Fig. 1A. Four genes, namely 2,5-DHP dioxygenase gene (hpo), N-formylmaleamate deformylase gene (nfo), maleamate amidase gene (ami) and maleate cis-trans isomerase gene (iso) in the nic2 cluster from strain S16, were cloned and expressed in Escherichia coli and all the related enzymes were characterized. The objective of this work is to demonstrate the unknown catabolism involved in the late steps of the nicotine degradation pathway.
Results
Identification of a gene cluster encoding the late pathway
The first fragment of a nic gene cluster (here designated as nic2) catalyzing the late steps of nicotine degradation was identified using in silico analysis of the P. putida S16 genome sequence, the nic gene cluster in KT2440 and the gene sequence of HspB. The entire 5.8-kb nic2 gene cluster was cloned into pMD18T. Resting cells of E. coli DH5α containing the pMD18T-nic2 construct degraded HSP completely within 9 h at 30°C and 37°C temperatures (data not shown). Fig. 1B shows the chromosomal nic2 gene cluster consisting of 6 open reading frames (ORFs). No sequences could be found by BLAST search, using the S16-nic cluster (hspB-ami part, 5,763 bp, GC content 47.9%) and nic cluster (iso-ami part, 3,233 bp, GC content 48.0%). The G+C content of the S16-nic2 cluster (47.9%) is considerably lower than the average for the P. putida S16 genome (62.3%). The 4 downstream enzymes catalyze steps 6-9 of the pathway (DHP to fumaric acid) (Fig. 1) and have orthologs in other bacteria, some of which are known to degrade nicotinic acid but not nicotine (Fig. 1B). The similarities among these orthologs are as low as 33% (indicated by the fading colors of the ORFs in Fig. 1). The dark cyan arrow indicates a nicotinic acid monooxygenase (6-hydroxy-nicotinate 3-monooxygenase, Hna), which has no ortholog in P. putida S16. The enzymatic activities of all four proteins of P. putida S16 have been verified. The presumptive ATG start codon was detected in nfo, hpo and ami, whereas the rare initiation codon GTG was found for iso. The Shine-Dalgarno sequences19 were identified in the upstream regions of the putative start codon of hpo ('GAGAGGCGCGATATG') and that of ami ('AAGGAGTATTATATG')19. The 4 genes are tightly linked, with no bases between the termination of iso and the start of nfo, 12 bases between the termination of nfo and the start of hpo and 38 bases between the termination of hpo and the start of ami. Non-quantitative reverse transcription polymerase chain reaction (RT-PCR) showed that the 4 genes appeared to be upregulated in 6 mM nicotine or 1 mM DHP addition separately, whereas no significant effects in the 4 genes expressions were observed with the individual addition of 1 mM NFM, 6 mM maleamic acid, or 6 mM maleic acid (Fig. S2A and Fig. S2B).
Cloning, purification and characterization of Hpo, Nfo, Ami and Iso
These enzymes were colorless, indicating that no tightly associated chromophores were present.
Hpo—The hpo gene was cloned from the gene cluster and over-expressed in E. coli BL21(DE3). Hpo was purified and showed an apparent molecular mass of 40 kDa which corresponds to the predicted gene product weight (Fig. 2A). The effect of pH on the spectrophotometric assay is shown in Fig. 2B. Optimum pH was 6.5 in phosphate buffer. The optimum temperature for the enzymatic reaction was approximately 20°C and was identified by measuring the initial reaction velocity for 1 min (Fig. 2C). Among the 10 metal salts tested, only Fe2+ (0.025 mM) showed markedly increased enzyme activity (Fig. 2D). The double reciprocal plots of initial velocity against DHP concentration were straight lines, indicating that Michaelis-Menten saturation kinetics were characteristic of the substrate. The calculated apparent Km for DHP was 0.168 μM, Vmax was 7.0 U mg−1 and kcat was 11.0 s−1. The product of the enzymatic reaction, N-formylmaleamate (NFM), was also detected by the API 4000 liquid chromatography-mass spectrometry (LC-MS) system (Applied Biosystems, Foster City, CA). A molecular ion at an m/z value of 112.2 corresponds to DHP according to positive mass spectrometry. Additionally, negative mass spectrometry experiments revealed an (M-H)- peak at m/z 142.1, corresponding to NFM, which is in agreement with that reported by Jimenez18 (Fig. S3).
Characterization of Hpo.
(A) SDS-PAGE. M, molecular size markers; lanes 1 and 2, cell extracts of expressed Hpo in E. coli BL21(DE3) expressing His6-Hpo; lane 3, purified His6-Hpo. (B) pH dependence of Hpo specific activity. (C) Effect of varying temperatures on specific Hpo activity. (D) Effect of the various metal salts on Hpo specific activity. CK, without metal salt; Ni, Ni2+; Co, Co2+; Ca, Ca2+; Cu, Cu2+; Mn, Mn2+; Zn, Zn2+; Mg, Mg2+; Mo, MoO42−; W, WO42−; Fe, Fe2+.
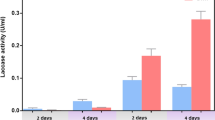
Nfo, Ami and Iso—The 29 kDa protein Nfo is consistent with the predicted product of the nfo gene (Fig. 3A). Because NFM was not commercially available, it was chemically synthesized and the product was confirmed by 1H nuclear magnetic resonance (NMR) (Fig. S4). The signals were assigned to the corresponding protons as indicated in the box (Fig. S4). Nfo activity was measured through its production of formate by a coupled reaction with nicotinamide adenine dinucleotide (NAD)-dependent formate dehydrogenase (Fdh) (Fig. 3B). Purified Ami (Fig. 3A) transformed maleamic acid to maleic acid with the production of NH4+ (not shown). NH4+ concentration was monitored by a decrease in the amount of nicotinamide adenine dinucleotide phosphate (NADPH) measured at 340 nm in a reaction buffer containing glutamate dehydrogenase (Gdh) (Fig. 3C). Iso was also expressed in E. coli BL21(DE3)/pET28a-iso and the 28-kDa enzyme was purified (Fig. 3D). Iso is similar to an isomerase from Ralstonia eutropha H16 (71% amino acid sequence homology) and to the Asp/Glu racemase of Serratia proteamaculans 568 (72% amino acid sequence homology). Iso activity was determined by high-performance liquid chromatography (HPLC) (Agilent 1200 series, Hewlett-Packard Corp., Santa Clara, CA, USA) analysis in the presence of a mercaptan. Purified Iso catalyzed the isomerization of maleate acid to fumaric acid (Fig. 3E). The apparent Km value of Iso was determined spectrophotometrically, monitoring maleate acid consumption at 290 nm. The assay was performed at 30°C in 50 mM Tris-HCl buffer (pH 8.4). The calculated apparent Km for maleate acid was 0.689 mM, Vmax was 9.4 U mg-1 proteins and kcat was 4.0 s−1 (Fig. 3F), respectively.
Characterization of Nfo, Ami and Iso.
(A) SDS-PAGE of Nfo and Ami. M, molecular size markers; lanes 1 and 2, cell extracts of E. coli BL21(DE3) expressing His6-Nfo; lane 3, purified His6-Nfo; lanes 4 and 5, cell extracts of E. coli BL21(DE3) expressing His6-Ami; 6, purified His6-Ami. (B) Nfo activity, a, complete reaction: 695 μl buffer + 5 μl NAD + 50 μl Fdh + 50 μl Nfo + 200 μl NFM; b, positive control containing HCOOH (1.65 mM) instead of Nfo and NFM; c, negative control without NFM; d, negative control without Nfo; e, negative control without Nfo and NAD. (C) Enzymatic activity of Ami, a, negative control without Ami; b, negative control, without maleamic acid and Ami; c, negative control without maleamic acid; d, complete reaction containing 480 μl buffer (50 mM Tris-HCl + 0.001 mol l−1 EDTA), 200 μl maleamic acid (0.034 mol l−1), 10 μl α-ketoglutaric acid (0.5 mol l−1), 10 μl NADPH (0.01 mol l−1), 100 μl Gdh (9.5 unit ml−1) and 200 μl amidase (0.106 mg protein ml−1); e, positive control, containing ammonia (30 μM NH4Cl) instead of maleamic acid and Ami. (D) SDS-PAGE of Iso. M, molecular size markers; lanes 1 and 2, cell extracts of expressed Iso in E. coli BL21(DE3); lanes 3 and 4, purified His6-tagged Iso. (E) Transformation of maleic acid by Iso. (F) Determination of Km of Iso.
Deletion of the hpo gene
The hpo gene was deleted separately and the related cell growth and resting cell reactions were all performed. The hpo gene deletion mutant could grow in nicotine medium and turn its color to saddle brown, whereas the color culture medium of the wild type strain S16 changed into pale green (Fig. 4A). Cells of hpo gene deletion mutant and wild type strain S16 were harvested in the mid-growth phase by centrifugation at 5,000 × g for 10 min at 4°C, washed three times with 0.02 M Tris-HCl buffer (pH 7) and suspended in the same buffer at OD600 nm (called resting cells) value of 7. Resting cells of both cell types have the ability to transform nicotine (Fig. 4B). However, resting cells of hpo gene deletion mutant could not degrade DHP, different from the wild-type strain S16 (Fig. 4C). These results demonstrate that enzyme Hpo is crucial for DHP transformation and nicotine degradation by the strain.
Nicotine transformation by P. putida S16 and P. putida S16 hpo::pk18mob.
(A) The liquid culture of P. putida S16 and P. putida S16 hpo::pk18mob in nicotine medium. The medium was cultured at 30°C for 16 h. (B) The concentration of nicotine in P. putida S16 and P. putida S16 hpo::pk18mob resting cell systems. The resting cell systems were cultured at 30°C; sampling was conducted at 0, 3 and 5 h. (C) The concentration of 2,5-DHP in P. putida S16 and P. putida S16 hpo::pk18mob resting cell systems. The resting cell systems were cultured at 30°C sampling was conducted at 0, 3 and 5 h.
Comparative genome analysis
Comparative genome analysis was performed using Mauve20 and genomic islands (GIs) were analyzed using the IslandViewer software21. Phylogenetic tree construction using MEGA 4.122 (containing genes hpo, nfo, ami and iso, Fig. 5A) showed that the gene cluster from S16 is most closely related to Octadecabacter antarcticus 238. The phylogenetic trees constructed with the neighbor joining (NJ) method for the individual proteins further refined the analysis, identifying similarities among these proteins (Fig. 5B). Hpo of P. putida S16 is closest to the ortholog from Rhizobium WSM1325, whereas Nfo and Ami are closest to O. antarcticus 238. Iso of P. putida S16 is in a deep branch of its own. Moreover, 29 GIs were identified in the complete genome of strain S16. The nic2 gene cluster is located within the largest GI (43,989 bp in length), which has a G+C content of 52.4% and codes for 8 transposases16 (Fig. S1). The GI containing the late pathway genes was not found in any of the 5 completely sequenced genome strains of P. putida namely P. putida KT2440, W619, F1, GB-1 and BIRD-1. The nicotinic acid degrading genes nicAB of P. putida KT244018 were found in the genomes of 4 other P. putida strains. However, the nicAB genes were not found in the strain S16 genome. Attempts to degrade nicotinic acid by resting cells of strain S16 did not result in any degradation after 24 h of incubation (data not shown).
Phylogenetic trees of the nic gene cluster and 4 related enzymes in late pathway of nicotine degradation.
The phylogenetic trees were constructed with the neighbor joining method (NJ). (A) Phylogenetic tree of nic clusters of different strains constructed using MEGA 4.1. (B) Phylogenetic trees of Hpo, Nfo, Ami and Iso of different strains constructed using MEGA 4.1.
Discussion
The analysis of genes and the biochemical characterization of nicotine catabolism enzymes serve as bases for rationally improving Pseudomonas strains for nicotine waste disposal2,4,23. The characterization of genes and gene products involved in nicotine degradation in Pseudomonas can lead to a full understanding of this catabolic activity. The identification of genes involved in the nicotine degradation pathway increases the possibility to clone related genes from other organisms, including noncultivable environmental samples. 6-Hydroxy-3-succinoylpyridine (HSP) and 2,5-dihydroxy-pyridine (DHP) in nicotine degradation are useful precursors for chemical synthesis10,13. Knowledge of nicotine degradation genes such as hpo and hspB15 can be adapted in detoxification programs for tobacco wastes and synthesis of useful products of pharmaceutical importance. These genes may also be useful for the modification or degradation of alkaloid substances and other heterocyclic aromatic compounds.
The complete proposed nicotine degradation pathway is presented in Fig. 1A. The discovered nic2 gene cluster encodes for 5 enzymes that convert HSP to fumaric acid, formic acid and ammonia (Fig. 1A). All of the enzymes were cloned and expressed in E. coli and their biochemical activities were confirmed. The nic2 gene cluster is separated by more than 30 kb of DNA from the nic1 gene cluster, which contains nicA and hspA13,14,15,16.
Several gene clusters in other microorganisms with similarity to nic2 in P. putida S16 were found in the databases (Fig. 1B). No reports on nicotine degradation in these strains have been published, except for strain S16. Gene sequence alignment showed that the 4 enzymes (Hpo, Nfo, Ami and Iso) showed variations (36%–69% amino acid homology), from their closest homologues in P. putida KT2440, which were not reported to degrade nicotine. The cluster (iso-ami part) shows a low level of homology to the partly syntenic gene clusters of other P. putida strains, KT2440, F1, GB-1 and W619, which encodes proteins of only 33%–72% amino acid homology (Fig. 1B). However, these strains may degrade nicotinic acid instead of nicotine and are very similar to each other (mostly > 95% amino acid homology, Fig. S5). The gene cluster that is highly similar to the cluster (iso-ami part) of P. putida S16 was from O. antarcticus 238, which has not been reported to be involved in nicotine or nicotinic acid degradation. The genes are syntenic and the corresponding proteins are 60%–81% homologous (Fig. 1B). The GC content of the 3,233-bp gene cluster (iso-ami part) is 48%. The genome of strain S16 consists of a single circular chromosome 5,984,790 bp in length, with a GC content of 62.3% and do not contain any plasmids16. In contrast, the GC contents of the nic clusters of the other P. putida strains are all approximately 64%. The relatively low GC content of the S16-nic2 gene cluster and the divergence of the amino acid sequences in Fig. 5 suggested that nic2 of P. putida S16 may have originated from a genus other than Pseudomonas.
Nicotinic acid degradation pathways have been characterized in several microorganisms such as Pseudomonas, Bacillus and Eubacterium17,18,24,25,26,27,28. Behrman and Stanier reported that nicotinic acid metabolism in Pseudomonas proceeds through the maleamate pathway, involving DHP, maleamate, maleate and fumarate as intermediates25. This is similar to the late pathway of nicotine degradation in strain S16. No ortholog of NicAB, the two-component nicotinic acid hydroxylase of P. putida KT244018, was found in the P. putida S16 genomic sequence, suggesting the inability of strain S16 to catabolize nicotinic acid.
The nicotine degradation genes may be useful for the genetic engineering of nicotine catabolism and for the production of specialist enzymes. The proposed reactions catalyzed by the 4 enzymes, 2,5-DHP dioxygenase (Hpo, EC 1.13.11.9), N-formylmaleamate deformylase (Nfo, EC 3.5.1.106), maleamate amidase (Ami, EC 3.5.1.107) and maleate cis-trans isomerase (Iso, EC 5.2.1.1), are particularly interesting. One of the key enzymes involved in nicotine degradation is the 2,5-DHP-oxidase (Hpo), which requires a ferrous ion for activation. Similar properties were observed in other dioxygenases that catalyze the cleavage of the aromatic ring18. This study proposes that the sensitivity to oxidizing agents is at least partially related to the weak binding of the catalytically active Fe2+ ion to Hpo. Hpo of P. putida S16 did not require an external reduced cofactor and contained no flavin or heme group. The Hpo enzymes (Fig. 1, orange arrows) from strain S16 and P. putida KT2440 catalyze the similar enzymatic reactions, although their protein sequences are only 43% homologous. Orthologs of Nfo, Ami and Iso from the nicotine degradation pathway were also characterized in this study. Both Nfo (deformylase) and Ami (amidase) belong to the hydrolase family, which act on carbon-nitrogen bonds of linear amides18,29. Nfo catalyzes the deformylation of NFM to release maleamic and formic acid30, whereas Ami catalyzes the hydrolysis of an amide29. Iso (maleate cis-trans isomerase) has orthologs in several maleate-assimilating bacteria and was shown to reversibly convert maleate to fumarate. Maleate cis-trans isomerases have received considerable research attention owing to its potential to serve as industrial catalysts, because fumarate is a substrate for aspartic acid formation31.
In summary, a gene cluster encoding the nicotine catabolic enzymes of the late steps in the nicotine degradation was discovered and biochemically characterized. Genomic analysis of strain S16 revealed that genes located in the largest genome island are crucial for nicotine degradation. This study has also identified genes that may be useful for the genetic engineering of nicotine catabolism and for the production of specialist enzymes.
Methods
Bacterial strains and growth conditions
E. coli BL21(DE3) was employed as a host for both plasmids and the expression strain, cultured in Lysogeny broth medium and maintained at 37°C. P. putida S16 was grown in Nic medium and incubated at 30°C9.
Chemicals and biochemicals
Fdh, Gdh, formate, α-ketoglutaric acid, maleamic acid, maleic acid, fumaric acid, dithiothreitol, NADPH and NAD, were obtained from Sigma-Aldrich (St. Louis, MO, USA). L-(-)-Nicotine (≥ 99% purity) was obtained from Fluka Chemie GmbH (Buchs Corp., Buchs, Switzerland).
NFM synthesis
The product was prepared as reported elsewhere with minor modifications27. Maleic anhydride (10 g) was heated to 80°C under a nitrogen atmosphere in a 250-ml round bottom flask equipped with a reflux condenser.Anhydrous formamide was added (10 ml) and the reaction mixture was stirred slowly with a magnetic stirrer for 10 h at 80°C. The product was analyzed by NMR spectroscopy.
Identification of genes in the late steps
A fragment containing the gene cluster involved in the late steps of nicotine degradation was identified through data mining for amino acid sequences of NicX, NicD, NicF and NicE of P. putida KT244018 with the genome sequence of P. putida S1616. Each gene in the fragment was subcloned into pET28a and functionally analyzed. DNA manipulation and transformation were performed according to standard procedures32. The primers employed in this study are listed in Table 1. Primers were designed to incorporate an NcoI site in the forward primer and an XhoI site in the reverse primer. The DNA fragments bearing hpo, nfo, ami and iso were amplified from genomic DNA using pfu polymerase (Tiangen, China) digested with NcoI and XhoI and ligated into the NcoI-XhoI sites of pET28a. The resulting plasmids were used to transform E. coli BL21(DE3). Cells harboring the recombinant plasmids were cultured in LB medium containing 100 mg liter-1 kanamycin and maintained at 37°C until the OD600 reading reached 0.6. IPTG was then added to a final concentration of 1 mM and the cultures were incubated for another 6 h at 30°C to express the proteins. His6-tagged Hpo, Nfo, Ami and Iso were purified using a column of Ni-NTA agarose (Qiagen).
Enzyme activities of Hpo, Nfo, Ami and Iso
Hpo activity was measured at 20°C using a UV-Vis 2550 spectrophotometer (Shimadzu) at 320 nm (ε320 = 5200 cm-1 M-1) as the previously reported17,18. The reaction mixture contained 0.02 mg ml-1 DHP, 0.025 mM Fe2+, 18.2 mM Tris-HCl buffer (pH 8.0) and 0.0125 mg protein ml-1 enzyme in a total volume of 1 ml. One unit of activity was defined as the amount of enzyme that catalyzed the disappearance of 1 μmol of substrate in 1 min.
Nfo and Ami activities were measured at 20°C, using the UV-Vis 2550 spectrophotometer at 340 nm. The Nfo assay mixture contained 695 μl Tris-HCl buffer (50 mM, pH 8.0), 5 μl NAD (0.33 M), 50 μl Fdh (9.2 unit ml-1), 200 μl NFM (286 μmol l-1) and 50 μl Nfo (0.57 mg protein ml-1). The Ami assay mixture contained 480 μl buffer (50 mM Tris-HCl + 0.001 mol l-1 EDTA), 200 μl maleamic acid (0.034 mol l-1), 10 μl α-ketoglutaric acid (0.5 mol l-1), 10 μl NADPH (0.01 mol l-1), 100 μl Gdh (9.5 unit ml-1) and 200 μl Ami (0.11 mg protein ml-1).
Iso activity was measured at 30°C, using the UV-Vis 2550 spectrophotometer at 290 nm as previously reported31. The Iso assay mixture contained 50 mM Tris-Cl (pH 8.4), 10 μl 1-thioglycerol (500 mM), an appropriate volume of maleate (1 M, pH 7) and 5 μl isomerase (10.1 mg protein ml-1) in a total volume of 1 ml.
Analytical techniques
DHP, NFM, maleamic acid and maleic acid were detected by HPLC and confirmed by direct-insertion mass spectra recorded on the API 4000 LC-MS system. MS analysis was performed in both negative and positive-ion turbo ion spray ionization mode.
Nonquantitative RT-PCR
RT-PCR procedure was followed as previously reported14. Total RNA was isolated from P. putida S16 by using a Total RNA kit I (Omega, USA). Contaminating DNA was treated with DNase I (RNAse-free; Fermentas, EU) at 1 U per 1 μg of total RNA for 30 min at 37°C. RT-PCR was performed in 50-μl reaction mixtures containing approximately 400 ng of total RNA and 20 pmol of each primer with a Prime ScriptTM one step RT-PCR kit (Takara, Japan). The amplification conditions employed for RT-PCR was as follows: 50°C for 30 min; 94°C for 2 min; 30 cycles of 94°C for 30 s; 57°C for hpo, nfo and ami, 68°C for iso and 72°C for 2 min. The primers employed in the amplification are listed in Table 1. RNA was used as a negative control to confirm the absence of DNA that may be contaminating the RNA preparations.
P. putida S16 mutant (hpo gene deletion)
To delete the hpo gene in strain S16 by single homologous recombination, internal fragments were amplified by PCR and cloned into the polylinker region of pK18mob (hpo -347-f: CAGTGAATTCAACCTCCTGAGCTTCTGGCCC; hpo -708-r: CTTAGTCGACTTCAAGACCGCCGCGAATATC). The pK18mob-hpo was transformed and the deletion mutant was screened as previously described15.
Nucleotide sequence accession number
The nucleotide sequence reported in the present study has been deposited in GenBank under accession number GQ857548.
References
Novotny, T. E. & Zhao, F. Consumption and production waste: another externality of tobacco use. Tob. Control 8, 75–80 (1999).
Brandsch, R. Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol. 69, 493–498 (2006).
Eberhardt, H. The Biological degradation of nicotine by nicotinophilic microorganisms. Beitr. Tabakforsch. Int. 16, 119–129 (1995).
Li, H. J., Li, X. M., Duan, Y. Q., Zhang, K. Q. & Yang, J. K. Biotransformation of nicotine by microbiology: the case of Pseudomonas spp. Appl. Microbiol. Biotechnol. 86, 11–17 (2010).
Ruan, A. D., Min, H., Peng, X. H. & Huang, Z. Isolation and characterization of Pseudomonas sp. strain HF-1, capable of degrading nicotine. Res. Microbiol. 156, 700–706 (2005).
Wada, E. & Yamasaki, K. Mechanism of microbial degradation of nicotine. Science 117, 152–153 (1953).
Wada, E. Microbial degradation of the tobacco alkaloids and some related compounds. Arch. Biochem. Biophys. 72, 145–162 (1957).
Wang, M. Z., Yang, C. Q., Min, H., Lv, Z. M. & Jia, X. Y. Bioaugmentation with the nicotine-degrading bacterium Pseudomonas sp. HF-1 in a sequencing batch reactor treating tobacco wastewater: Degradation study and analysis of its mechanisms. Water Res. 43, 4187–4196 (2009).
Wang, S. N. et al. Biodegradation and detoxification of nicotine in tobacco solid waste by a Pseudomonas sp. Biotechnol. Lett. 26, 1493–1496 (2004).
Wang, S. N., Xu, P., Tang, H. Z., Meng, J., Liu, X. L. & Ma, C. Q. ‘Green’ route to 6-hydroxy-3-succinoyl-pyridine from (S)-nicotine of tobacco waste by whole cells of a Pseudomonas sp. Environ. Sci. Technol. 39, 6877–6880 (2005).
Zhong, W. H. et al. Degradation of nicotine in tobacco waste extract by newly isolated Pseudomonas sp. ZUTSKD. Bioresource Technol. 101, 6935–6941 (2010).
Wang, S. N., Liu, Z., Tang, H. Z., Meng, J. & Xu, P. Characterization of environmentally friendly nicotine degradation by Pseudomonas putida biotype a strain S16. Microbiology-SGM 153, 1556–1565 (2007).
Tang, H. Z. et al. A novel gene, encoding 6-hydroxy-3-succinoylpyridine hydroxylase, involved in nicotine degradation by Pseudomonas putida strain S16. Appl. Environ. Microbiol. 74, 1567–1574 (2008).
Tang, H. Z. et al. Novel nicotine oxidoreductase-encoding gene involved in nicotine degradation by Pseudomonas putida strain S16. Appl. Environ. Microbiol. 75, 772–778 (2009).
Tang, H. Z. et al. A Novel NADH-dependent and FAD-containing hydroxylase is crucial for nicotine degradation by Pseudomonas putida. J. Biol. Chem. 286, 39179–39187 (2011).
Yu, H. et al. Complete genome sequence of nicotine-degrading Pseudomonas putida strain S16. J. Bacteriol. 193, 5541–5542 (2011).
Gauthier, J. J. & Rittenberg, S. C. The metabolism of nicotinic acid. I. Purification and properties of 2, 5-dihydroxypyridine oxygenase from Pseudomonas putida N-9. J. Biol. Chem. 246, 3737–3742 (1971a)
Jimenez, J. I. et al. Deciphering the genetic determinants for aerobic nicotinic acid degradation: The nic cluster from Pseudomonas putida KT2440. Proc. Natl. Acad. Sci. USA 105, 11329–11334 (2008).
Shine, J. & Dalgarno, L. Determinant of cistron specificity in bacterial ribosomes. Nature 254, 34–38 (1975).
Darling, A. C., Mau, B., Blattner, F. R. & Perna, N. T. Mauve:multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004).
Langille, M. G. & Brinkman, F. S. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25, 664–665 (2009).
Kumar, S., Nei, M., Dudley, J. & Tamura, K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9, 299–306 (2008).
Civilini, M., Domenis, C., Sebastianutto, N. & Bertoldi,. Md. Nicotine decontamination of tobacco agro-industrial waste and its degradation by micro-organisms. Waste Manage. Res. 15, 349–358 (1997).
Alhapel, A. et al. Molecular and functional analysis of nicotinate catabolism in Eubacterium barkeri. Proc. Natl. Acad. Sci. USA 103, 12341–12346 (2006).
Behrman, E. J. & Stanier, R. Y. The bacterial oxidation of nicotinic acid. J. Biol. Chem. 228, 923–945 (1957).
Ensign, J. C. & Rittenberg, S. C. The pathway of nicotinic acid oxidation by a Bacillus species. J. Biol. Chem. 239, 2285–2291 (1964).
Gauthier, J. J. & Rittenberg, S. C. The metabolism of nicotinic acid. II. 2, 5-dihydroxypyridine oxidation, product formation and oxygen 18 incorporation. J. Biol. Chem. 246, 3743–3748 (1971b
Nakano, H. et al. Purification, characterization and gene cloning of 6-hydroxynicotinate 3-monooxygenase from Pseudomonas fluorescens TN5. Eur. J. Biochem. 260, 120–126 (1999).
Bray, H. G., James, S. P., Raffan, I. M., Ryman, B. E. & Thorpe, W. V. The fate of certain organic acids and amides in the rabbit. 7. An amidase of rabbit liver. Biochem. J. 44, 618–625 (1949).
Mazel, D., Pochet, S. & Marliere, P. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO. J. 13, 914–923 (1994).
Scher, W. & Jakoby, W. B. Maleate isomerase. J. Biol. Chem. 244, 1878-1882 (1969).
Sambrook, J. & Russell, D. W. Molecular cloning: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory (2001).
Acknowledgements
This work was supported in part by Chinese National Natural Science Foundation (30900042 and 30821005). We also acknowledge the “Chen Guang” project from the Shanghai Municipal Education Commission and the Shanghai Education Development Foundation (10CG10) and the “Chen Xing” project from Shanghai Jiaotong University.
Author information
Authors and Affiliations
Contributions
P.X. and H.T. conceived and designed the project and experiments. H.T., Y.Y., L.W., H.Y. and Y.R. performed the experiments. P.X. and G.W. analyzed the data. H.T. and P.X. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
supporting information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Tang, H., Yao, Y., Wang, L. et al. Genomic analysis of Pseudomonas putida: genes in a genome island are crucial for nicotine degradation. Sci Rep 2, 377 (2012). https://doi.org/10.1038/srep00377
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00377
This article is cited by
-
Nicotine metabolism pathway in bacteria: mechanism, modification, and application
Applied Microbiology and Biotechnology (2022)
-
Structure-guided insights into heterocyclic ring-cleavage catalysis of the non-heme Fe (II) dioxygenase NicX
Nature Communications (2021)
-
Influence of microbiota and metabolites on the quality of tobacco during fermentation
BMC Microbiology (2020)
-
Isolation of a 3-hydroxypyridine degrading bacterium, Agrobacterium sp. DW-1, and its proposed degradation pathway
AMB Express (2019)
-
Co-occurrence of functional modules derived from nicotine-degrading gene clusters confers additive effects in Pseudomonas sp. JY-Q
Applied Microbiology and Biotechnology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.