Abstract
We describe a site-specific recombination-based tandem assembly (SSRTA) method for reconstruction of biological parts in synthetic biology. The system was catalyzed by Streptomyces phage φBT1 integrase, which belongs to the large serine recombinase subfamily. This one-step approach was efficient and accurate and able to join multiple DNA molecules in vitro in a defined order. Thus, it could have applications in constructing metabolic pathways and genetic networks.
Similar content being viewed by others
Introduction
Synthetic biology is more ambitious than conventional genetic engineering and aims to design and reconstruct biological systems or even entire bacterial genomes. An essential technique in synthetic biology is the physical assemble of multiple small DNA fragments into large constructs with a defined order and orientation1,2. Traditional methods rely on restriction enzyme digestion followed by ligation and while this approach works well for the insertion of a single DNA sequence into a vector, as well as being time-consuming, it is often hard to find enough distinct restriction sites for the cloning of multiple DNA fragments1. Two techniques, BioBrick™ developed at MIT3 and BglBrick developed at UC Berkeley4, standardized the process of DNA assembly by using standard restriction sites. Biological parts were flanked by four restriction sites (two on each side) and the assembly products of the first round can subsequently be used in the next round, thus permitting the assembly of multiple parts.
The BioBrick2 method is limited by a number of ‘forbidden sites’. This has resulted in the development of a series of ligation-independent cloning (LIC) strategies that offered sequence-independent assembly. These include SLIC (sequence and ligase independent cloning)5, In-Fusion™ (Clontech) technologies6, the ‘Gibson’ isothermal assembly method7 and CPEC (circular polymerase extension cloning)8,9. These methods are dependent on overlapping homologous sequences at the ends of the DNA fragments1,2. SLIC, In-Fusion™ and the Gibson isothermal assembly method use a mechanism called ‘chew back and anneal’2,7,10,11, while CPEC relies on annealing of complimentary DNA regions and a single high-fidelity PCR cycle9. Using the isothermal assembly method, Gibson and his colleagues successfully assembled three 5-kb DNA molecules using fragments of only 40 bp overlaps. This method was also used to assemble a 900-kb molecule in vitro7. The Gibson method was also effective in gene synthesis and an entire 16.3-kilobase mouse mitochondrial genome was synthesized through three rounds of assembly from 600 overlapping 60-mers12.
Several competing approaches using type IIs restriction enzymes have also been developed, such as Golden Gate cloning13,14 and Pairwise Selection Assembly (PSA)15. Since type IIs restriction enzymes cut outside of their recognition sequence, through proper design of the cleavage sites this approach is scarless14. Using Golden Gate cloning, Weber et al13 assembled a 33-kb DNA molecule from 44 individual parts in three steps. Blake et al15 used PSA to reconstruct a completely synthetic 91-kb molecule in six rounds of assembly. In general, assembly of DNA fragments occurs on three scales – ‘parts to genes’, ‘genes to pathway’ and ‘pathways to genomes’2 – and each assembly strategy described above has its own advantages and disadvantages at different scales1,2.
We have developed an alternative strategy for the assembly of multiple DNA segments in one reaction based on φBT1 integrase-mediated recombination in vitro, termed site-specific recombination-based tandem assembly (SSRTA). We previously established a site-specific recombination system based on Streptomyces phage φBT1 integrase-mediated integration in vitro16 and identified 16 pairs of non-compatible attB and attP recombination sites17. The φBT1 integration system is highly efficient and accurate because no recombination can occur between att sites with different central di-nucleotides17. The SSRTA method is sequence-independent and excellent for the combinatorial assembly of multiple biological parts in a defined order, even with those with high GC contents (70%). The utility of SSRTA was demonstrated in the assembly of an entire epothilone biosynthetic gene cluster (62.4 kb) from ten individual parts.
Results
The recombination efficiencies of φBT1 integrase-catalyzed reactions between attB and attP sites in vitro were over 90% in our previous studies16,17 and could therefore be used for establishing a robust DNA assembly approach. This system contains 16 pairs of mutated recombination sites, the purified φBT1 integrase and a simple buffer (see Materials and Methods). Once the target DNA modules have been flanked by a pair of non-compatible att sites, multiple fragments can be assembled in one reaction simultaneously at 30°C in vitro (Fig. 1). Because site-specific recombination occurs by precise breakage-joining events and do not involve any DNA synthesis or loss, this method is highly accurate and no errors are introduced after assembly.
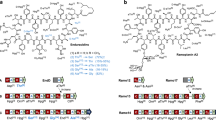
Site-specific recombination-based tandem assembly in vitro.
Multiple DNA modules were flanked by pairs of non-compatible recombination sites of the φBT1 integration system. A series of mutated attB sites were placed upstream of each module and mutated attP sites were located downstream. After incubation of all the DNA modules with φBT1 integrase, tandemly assembled products are produced in a one-step reaction. Differently coloured arrows represented different pairs of recombination sites.
We selected the epothilone biosynthetic gene cluster to test this SSRTA method. Epothilone polyketides are promising anti-cancer drugs with remarkable microtubule-stabilizing activity. The total length of the gene cluster is nearly 56 kb and consists of six open reading frames (epoA to epoF) and ten modules, including one loading module (LM), one non-ribosomal peptide synthetase (NRPS) module and eight polyketide synthase (PKS) modules. Modules 3 to 6 form a whole ORF defined as epoD, while modules 7 and 8 form epoE18 (see Supplemental Fig. S1). EpoK, a cytochrome P450 involved in epoxidation of epothilones C and D to A and B19, was not included in this work. The purified genomic DNA of the epothilone-producing myxobacterial strain Sorangium cellulosum So0157-2 was used as the template for PCR amplification of the modules20. Due to the high GC content (69.5%) of the epothilone biosynthetic gene cluster, we adopted an ‘entry clone’ strategy to place individual modules into a series of vectors containing the appropriate pairs of recombination sites beforehand, instead of engineering the recombination sites into the 5′-terminus of PCR primers.
We constructed a series of ‘entry vectors’ with the apramycin resistance gene (aac(3)IV) flanked by a pair of recombination sites. We also put two XcmI recognition sites close to the recombination sites. After the vectors were digested with XcmI, the target DNA modules were inserted into the ‘entry vectors’ through TA cloning (Fig. 2a). The whole epothilone biosynthetic gene cluster was divided into small parts to facilitate amplification by high-fidelity PCR (PrimerSTAR™ HS DNA polymerase, Takara) (Supplemental Fig. S1). Through PCR amplification and sub-cloning, we constructed nine independent ‘entry clones’ which contained all the essential genes for epothilones production. The ribosomal binding site sequences were fused in the 5′-termus of each open reading frame (ORF) by PCR amplification (Supplemental Fig. S1). After linearization by restriction enzymes, these ‘entry clones’ were ready for tandem assembly in vitro. It should be noted that the genes in each entry clone can be replaced by any other desired module. The details of the construction process are described in Materials and Methods, Supplemental Fig. S1 and Tables S1 and S2. Plasmid pZLE10 was designed to propagate the circular assembly products in Escherichia coli and then to integrate into the Streptomyces genome for epothilone production (Fig. 2b).
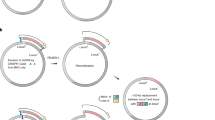
Strategy for assembling the epothilone biosynthetic gene cluster by SSRTA.
(a) Map of seven ‘entry vectors’ used in this study. Arrows in pink, green, dark green, red, lime, orange and blue represent pairs of recombination sites numbered 0, 6, 13, 7, 12, 3 and 15, respectively. (b) Map of plasmid pZLE10. Details are available in the Online Methods and Supplementary Table 2. (c) Schematic of the tandem assembly of pZLE10-epoD. Five groups of recombination reactions occur simultaneously, between attB0/attP0 , attB6/attP6 , attB13/attP13 , attB3/attP3 and attB15/attP15 . (d) Schematic of pZLE10-epo assembly. Seven groups of recombination reactions take place, between attB0/attP0 , attB6/attP6 , attB13/attP13 , attB7/attP7 , attB12/attP12 , attB3/attP3 and attB15/attP15 .
The construction process was divided into two steps: first we combined epoD DNA sequences from five individual clones and removed the scar sequences inside the ORF. Then we assembled all the epothlione biosynthetic genes from seven individual clones in one reaction. First we tested the ability to combine epoD from four ‘entry clones’ (Fig. 2c). The plasmids pTA0006-M3-aphII, pTA0613-M4, pTA1303-M5, pTA0315-M6 and pZLE10 were linearized (Supplemental Fig. S2) and incubated with φBT1 integrase overnight (or over 8 hrs) at 30°C. As shown in Fig. 3a, multiple assembly products were obtained (the correct, complete assembly product was a 29.8-kb plasmid). The in vitro reaction products were transformed into E. coli strain DH10B by electroporation. After selection with apramycin and neomycin, resistant clones were isolated and the plasmid DNA was extracted. PCR analysis of the linker regions between modules and restriction enzyme digestion indicated that the five segments had assembled correctly (Fig. 3b, c, Supplemental Fig. S3 and S4). We next explored the possibility of combining seven segments in a one-step reaction to generate pZLE10-epo (Fig. 2d, Supplemental Fig. S2 and S4). As shown in Fig. 3d-f, we obtained the assembled plasmid (62.4 kb) successfully. Thus, this tandem assembly system can be used to assemble at least seven DNA segments into a circular molecule in vitro.
Identification of the assembly products.
(a) Analysis of assembled pZLE10-epoD by pulsed-field gel electrophoresis. The sizes of the substrates were 6316 bp, 6610 bp, 7417 bp, 5178 bp and 7115 bp. The final assembly product was a 29-kb plasmid. (b) Identification of pZLE10-epoD by PCR of the linker regions. The predicted sizes were 322 bp (vm3), 1615 bp (m3m4), 177 bp (m4m5), 249 bp (m5m6) and 468 bp (m6v). (c) ApaI (lane 1), HindIII & NheI (lane 2) digests of pZLE10-epoD. (d) Analysis of assembled pZLE10-epo by pulsed-field gel electrophoresis. The sizes of substrates were 4447 bp, 5749 bp, 7019 bp, 24857 bp, 11568 bp, 8840 bp and 7115 bp and the final assembly product was a 62.4-kb plasmid. (e) Identification of pZLE10-epo by PCR of the linker regions. The predicted sizes were 467 bp (vm0), 420 bp (m0m1), 420 bp (m1m2), 386 bp (m2m3), 177 bp (m4m5), 249bp (m5m6), 277 bp (m6m7), 306 bp (m8m9) and 411 bp (m9v). (f) HindIII (lane 1) and NdeI (lane 2) digests of pZLE10-epo.
Discussion
DNA assembly provides a means for extensively changing the genetics of entire biological pathways and cells. This may be useful for creating organisms capable of more than overproduce individual proteins – a common goal of cut-and-paste cloning techniques. Several successful approaches have been described that achieve this aim, such as BioBrick standard assembly3, the ‘Gibson’ isothermal assembly method7,12 and the Golden Gate assembly method14. In this manuscript we have demonstrated the utility of the SSRTA method by reconstructing the epothilone biosynthetic gene cluster. Seven independent DNA fragments were assembled into a circular plasmid (62.4 kb) through a one-step in vitro incubation. An ideal assembly method should have no forbidden sites, would be suitable for combinatorial assembly of different parts from standard modules and importantly, would allow assembly of the parts in a defined order2. The SSRTA method described here is based on site-specific recombination between a series of non-compatible attB and attP sites and so concerns about forbidden sites are not relevant. It is notable that the epothilone biosynthetic gene cluster contains ten direct repeat sequences larger than 100 bp, including a 554-bp direct repeat. Many antibiotic biosynthetic gene clusters contains such direct repeats21,22. Homology-based assembly methods may not be appropriate in these cases, but SSRAT has proved successful. Furthermore, each ‘entry vector’ could be used to build libraries of biological parts. These parts could be reconstructed in a defined order through combinatorial assembly (Supplemental Fig. S5). PCR products could also be directly used in this SSRTA method. Details of the pairs of non-compatible recombination sites and the sequences of PCR primers are described in Supplemental Fig. S6. The shortcoming of the SSRTA method is that it introduces scar sequences (attR, 42 bp) between modules after assembly, although scar sequences between genes may not be problematic2.
Synthetic biology holds promise in the development of cheaper drugs and ‘green’ biofuels, efficient environmental remediation and targeted therapies for diseases23,24. This SSRTA method provides an alternative way to rebuild genetic pathways and networks based on a phage-encoded large serine recombinase. Since nearly ten of these recombinases have been biochemically characterized25,26, this method could be widely adopted in other systems. This would also allow applications that combine multiple SSRTAs.
Methods
System design and construction of the ‘entry vectors’
A major feature of this SSRTA system is the use pairs of non-compatible recombination sites. Non-compatible sites cannot recombine with each other and the stringent control that this offers is the main strength of the system (Fig. 1). To assemble the full-length epothilone biosynthetic gene cluster (see supplemental Fig. S1) from six independent clones, seven pairs of attB/attP sites are needed. We chose pairs 0, 3, 6, 7, 12, 13 and 15, the sequences of which are given in our previous report17. These recombination sites were used to construct seven ‘entry vectors’, named pTA0006, pTA0613, pTA1307, pTA0712, pTA1203, pTA0315 and pTA1303 (Fig. 2a). pTA0006 was generated using primers PT1 (containing attB0 ) and PTF (containing attP6 ) to amplify the apramycin-resistance gene (aac(3)IV) and the PCR product was inserted into pMD19-T by TA cloning. Similarly, primers PTB6 (containing attB6 ) and PTP13 (containing attP13 ) were used to construct pTA0613; primers PTB13 (containing attB13 ) and PTP7 (containing attP7 ) were used to construct pTA1307; primers PTB7 (containing attB7 ) and PTP12 (containing attP12 ) were used to construct pTA0712; primers PTB12 (containing attB12 ) and PTP3 (containing attP3 ) were used to construct pTA1203; primers PTB3 (containing attB3 ) and PTP15 (containing attP15 ) were used to construct pTA0315; and primers PTB13 (containing attB13 ) and PTP3 (containing attP3 ) were used to construct pTA1303 (Supplemental Table S1 and S2). The apramycin-resistance gene was cloned into the ‘entry vectors’ in the same orientation in each case, as shown in Fig. 2a.
Construction of the ‘entry clones’ and destination vector
The complete epothilone biosynthetic gene cluster was divided into 17 parts for PCR amplification (Supplemental Fig. S1). To clone epoA, a 4289-bp DNA fragment was amplified using primers 00a and 00b and then inserted into pTA0006 by TA cloning to generate pTA0006-M0; for epoB, a 4256-bp DNA fragment was amplified using primers 01a and 01b and then inserted into pTA0613 to generate pTA0613-M1; for epoC, a 5525-bp DNA fragment was amplified using primers 02a and 02b and then inserted into pTA1307 to generate pTA1307-M2; for module 3, a 4766-bp DNA fragment was amplified using primers 03a and 03b and then inserted into pTA0006 to generate pTA0006-M3. Module 4 was divided into two parts, M4a (primers 04a/ZE05, 3252 bp) and M4b (primers ZE06/04b, 3220 bp); these were amplified and cloned into pTA0613 separately and then combined into one plasmid to generate pTA0613-M4 by enzyme digestion and ligation. Modules 5, 6, 7 and 8 were each divided into two parts. M5a (primers 05a/ZE07, 3607 bp) and M5b (primers ZE08/05b, 2767 bp) were PCR amplified and cloned into pTA1303 separately and then combined into one plasmid to generate pTA1303-M5. M6a (primers 06a/ZE09, 2490 bp) and M6b (primers ZE10/06b, 2611 bp) were PCR amplified and cloned into pTA0315 separately and then combined into one plasmid to generate pTA0315-M6. M7a (primers 07a/ZE11, 3506 bp) and M7b (primers ZE12/07b, 2683 bp) were PCR amplified and cloned into pTA1203 separately. M8a (primers 08a/ZE13, 2649 bp) and M8b (primers ZE14/08b, 3188 bp) were PCR amplified and cloned into pTA1203 separately. Plasmids containing genes from modules 7 and 8 were combined together by ligation after enzyme digestion to generate pTA1203-epoE. Module 9 was divided into three parts, M9a (primers 09a/ZE15, 3088 bp), M9b (primers ZE16/ZE19, 2229 bp) and M9c (primers ZE20/09b, 2443 bp); they were PCR amplified and cloned into pTA0315 separately and then combined into one plasmid to generate pTA0315-M9. The correct orientation of all inserts in the ‘entry clones’ was confirmed. To facilitate selection in the tandem assembly process, a PCR fragment (primers Oxj128R/Oxj129, 1338 bp) containing the aphII gene (conferring resistance to kanamycin and neomycin) was inserted into pTA0006-M3 through SpeI digestion and ligation (see Supplementary Table S1 and S2).
The destination vector pZLE10 used for integration and expression of the assembled gene cluster in Streptomyces was constructed as follows. A PCR fragment (primers ZE03/ZE04, 1956 bp) containing the redP promoter, the terminator from pIJ602127, the chloramphenicol resisitance gene and the attP0 and attB15 sites were inserted into the pMD-19T vector (Takara) to generate an intermediate plasmid. This was digested with XbaI and BglII to release a 1962-bp fragment. Plasmid T-Bxbatt1-Bxbatt2 (B. Zhang, L. Zhang, R. Dai, M. Yu, G. Zhao and X. Ding, manuscript in preparation) was digested with NheI and BglII to release a 4441-bp fragment containing apramycin and ampicillin resistance genes and the replication origin of pUC19 and p15A. These two fragments were ligated to generate pZLE12. pZLE12 was then digested with XbaI to release a 3713-bp fragment and plasmid pSET152 was digested with NheI and XbaI to release a 3402-bp fragment; these two fragments were ligated to generate pZLE10 (see Supplemental Table S2).
Tandem assembly of epoD and the complete epothilone biosynthetic gene cluster
The enzymes used to linearize the ‘entry clones’ for in vitro assembly procedures are described in Supplemental Fig. S2. DNA segments under 10 kb were isolated by 0.8% agarose gel electrophoresis in 1×TAE buffer and purified using an agarose gel DNA extraction kit (Generay); segments larger than 10 kb were purified by phenol-chloroform extraction and ethanol precipitation. For pZLE10-epoD assembly, the five DNA segments containing modules 3, 4, 5 and 6 and pZLE10 were incubated with φBT1 integrase overnight (or over 8 hrs) at 30°C. The buffer contained 10 mM Tris-HCl, 100 mM KCl, 50 mM NaCl, 2 mM EDTA and 1 mM DTT. Heterologous expression, purification and preservation of φBT1 integrase were described in our previous work16. The tandemly assembled products were purified by phenol-chloroform extraction and ethanol precipitation and analysis by pulsed-field gel electrophoresis (PFGE) performed on 1% agrose gels (Biolab) in 0.5×TBE buffer, with the following parameters: switch time, 1–6 seconds; run time, 11 hours; angle, 120°; and voltage gradient, 6 V/cm. Recombination events were detected by PCR analysis. The assembled products were transformed into E. coli strain DH10B by electroporation (25 μF capacitance, 1.8 kV/mm and 200 ohm resistance). Transformants were selected with apramycin and neomycin. The plasmid from surviving clones were identified by PCR using primers ES53/54, then isolated and purified. The plasmid was identified by further PCR reactions of the linker regions between modules and by restriction digestion (Fig. 3b, c and Supplemental Fig. S3). Correct clones were propagated for the next round of assembly. For assembly of the total epothilone biosynthetic gene cluster, the seven DNA segments containing epoA, epoB, epoC, epoD, epoE, epoF and pZLE10 were incubated with φBT1 integrase and the subsequent steps were as described above.
References
Hillson, N. J. in Design and Analysis of Biomolecular Circuits: Engineering Approaches to Systems and Synthetic Biology 295–314 (Springer-Verlag, Berlin, 2011).
Ellis, T., Adie, T. & Baldwin, G. S. DNA assembly for synthetic biology: from parts to pathways and beyond. Integr. Biol. 3, 109–118 (2011).
Smolke, C. D. Building outside of the box: iGEM and the BioBricks Foundation. Nat. Biotechnol. 27, 1099–1102 (2009).
Anderson, J. . C.., D. J., Leguia, M., Wu, G. C., Goler, J. A., Arkin, A. P., Keasling, J. D. Bglbricks: a flexible standard for biological part assembly. J. Biol. Eng. 4(1):1 (2010).
Elledge, S. J. & Li, M. Z. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 (2007).
Berrow, N. S. et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res 35, e45 (2007).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Tian, J. D. & Quan, J. Y. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 6, 242–251 (2011).
Quan, J. Y. & Tian, J. D. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 4 (2009).
Freeman, G. J., Zhu, B. G., Cai, G. F. & Hall, E. O. In-Fusion™ assembly: seamless engineering of multidomain fusion proteins, modular vectors and mutations. BioTechniques 43, 356–359 (2007).
Sleight, S. C., Bartley, B. A., Lieviant, J. A. & Sauro, H. M. In-Fusion BioBrick assembly and re-engineering. Nucleic Acids Res. 38, 2624–2636 (2010).
Gibson, D. G., Smith, H. O., Hutchison, C. A., Venter, J. C. & Merryman, C. Chemical synthesis of the mouse mitochondrial genome. Nat. Methods 7, 901–905 (2010).
Weber, E., Engler, C., Gruetzner, R., Werner, S. & Marillonnet, S. A modular cloning system for standardized assembly of multigene constructs. PLoS ONE 6 (2011).
Engler, C., Kandzia, R. & Marillonnet, S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3 (2008).
Blake, W. J. et al. Pairwise selection assembly for sequence-independent construction of long-length DNA. Nucleic Acids Res 38, 2594–2602 (2010).
Zhang, L., Ou, X. J., Zhao, G. P. & Ding, X. M. Highly efficient in vitro site-specific recombination system based on Streptomyces phage φBT1 integrase. J. Bacteriol. 190, 6392–6397 (2008).
Zhang, L. et al. DNA cleavage is independent of synapsis during Streptomyces phage φBT1 integrase-mediated site-specific recombination. J. Mol. Cell Biol. 2, 264–275 (2010).
Tang, L. et al. Cloning and heterologous expression of the epothilone gene cluster. Science 287, 640–642 (2000).
Ogura, H. et al. EpoK, a cytochrome P450 involved in biosynthesis of the anticancer agents epothilones A and B: substrate-mediated rescue of a P450 enzyme. Biochemistry 43, 14712–14721 (2004).
Gong, G. L. et al. Mutation and a high-throughput screening method for improving the production of Epothilones of Sorangium . J. Ind. Microbiol. Biotechnol. 34, 615–623 (2007).
Shen, B., Du, L. C., Sanchez, C., Chen, M. & Edwards, D. J. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem. Biol. 7, 623–642 (2000).
Leadlay, P. F. et al. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 25, 447–453 (2007).
Benner, S. A. & Sismour, A. M. Synthetic biology. Nat. Rev. Genet. 6, 533–543 (2005).
Khalil, A. S. & Collins, J. J. Synthetic biology: applications come of age. Nat. Rev. Genet. 11, 367–379 (2010).
Zhang, L., Zhao, G. P. & Ding, X. M. Site-specific recombination systems: mechanisms and applications. SCIENTIA SINICA Vitae 40, 1090–1111 (2010).
Grindley, N. D. F., Whiteson, K. L. & Rice, P. A. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 75, 567–605 (2006).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics (The John Innes Foundation, Norwich, 2000).
Acknowledgements
We thank Prof. Yuezhong Li (Shandong University, China) for providing the Sorangium cellulosum strain So0157-2. This work was supported by grants from the National High Technology Research and Development Program of China (863 Program) (grant number 2007AA021206), National Basic Research Program of China (973 Program) (grant number 2012CB721102) and the National Natural Science Foundation of China (grant numbers 30600009, 30830002).
Author information
Authors and Affiliations
Contributions
L.Z., G.-P.Z. and X.-M.D. designed research; L.Z. performed research; L.Z. and X.-M.D. analyzed data; and L.Z., G.-P. Z. and X.-M.D. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Zhang, L., Zhao, G. & Ding, X. Tandem assembly of the epothilone biosynthetic gene cluster by in vitro site-specific recombination. Sci Rep 1, 141 (2011). https://doi.org/10.1038/srep00141
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00141
This article is cited by
-
Influence of domestication on specialized metabolic pathways in fruit crops
Planta (2021)
-
In Vitro Seamless Stack Enzymatic Assembly of DNA Molecules Based on a Strategy Involving Splicing of Restriction Sites
Scientific Reports (2017)
-
A strategy for enhanced circular DNA construction efficiency based on DNA cyclization after microbial transformation
Microbial Cell Factories (2015)
-
Direct cloning and heterologous expression of the salinomycin biosynthetic gene cluster from Streptomyces albus DSM41398 in Streptomyces coelicolor A3(2)
Scientific Reports (2015)
-
Bricks and blueprints: methods and standards for DNA assembly
Nature Reviews Molecular Cell Biology (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.