Abstract
Concurrent regional and global environmental changes are affecting freshwater ecosystems. Decadal-scale data on lake ecosystems that can describe processes affected by these changes are important as multiple stressors often interact to alter the trajectory of key ecological phenomena in complex ways. Due to the practical challenges associated with long-term data collections, the majority of existing long-term data sets focus on only a small number of lakes or few response variables. Here we present physical, chemical, and biological data from 28 lakes in the Adirondack Mountains of northern New York State. These data span the period from 1994–2012 and harmonize multiple open and as-yet unpublished data sources. The dataset creation is reproducible and transparent; R code and all original files used to create the dataset are provided in an appendix. This dataset will be useful for examining ecological change in lakes undergoing multiple stressors.
Design Type(s) | observation design • time series design • data integration objective |
Measurement Type(s) | geographic location • Physical Phenomenon or Property • Inorganic Chemistry • planktonic material • weather |
Technology Type(s) | digital curation • data acquisition system • Analytical Chemistry • light microscopy |
Factor Type(s) | |
Sample Characteristic(s) | Adirondack Park • freshwater lake biome |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
Freshwater lakes are changing in complex ways, with multiple long-term environmental stressors interacting to form novel conditions in aquatic ecosystems. Most lakes globally are undergoing temperature warming in response to climate change1,2. Many lakes also face concurrent stressors such as acidification and subsequent recovery3–5, browning6, eutrophication7,8, invasive species9,10, and/or increased extraction for drinking water or irrigation11. While some stressors act at global scales (e.g., climate change), many stressors are local or regional. For example, many lakes in the northeastern U.S. and northern Europe were strongly acidified in past decades due to sulfur and nitrogen deposition from emissions from fossil fuel combustion and agricultural activity12,13 and have begun recovering since then in response to regulated decreases in emissions4,5,14. In agricultural areas, nutrient use and consequent eutrophication continue to result in water-quality issues such as anoxia15,16 and harmful algal blooms17.
Long-term data are critical to understanding and predicting the effects of ecosystem stressors that may act on decadal to multi-decadal scales18. Moreover, ecosystems can experience multiple, concurrent stressors. Understanding the effects of multiple, concurrent stressors is a critical need as disturbance regimes may interact to alter the trajectory of important biological and biogeochemical phenomena in complex ways. Regional and global-scale changes that occur simultaneously highlight the critical need for quality, long-term data on lake ecosystems that describe processes, interactions, and responses to multiple stressors. A number of existing long-term limnological datasets have been used to understand some aspects of long-term ecosystem change. For example, a recent analysis of eleven diverse lakes in the North Temperate Lakes (NTL) Long-Term Ecological Research (LTER) site has shown seasonal heterogeneity of water temperature warming in response to regional climate change19. Long-term monitoring of lakes in Europe and the United States have observed changes in water clarity20 and warming surface temperatures21 and a recently published 80-year data record showed the influence of re-forestation on long-term browning of Swedish lakes22. Such long-term datasets have formed the foundation of our modern understanding of limnological change. However, due to the many challenges associated with long-term data collections, the majority of long-term data sets focus on only a small number of lakes or response variables, but rarely both.
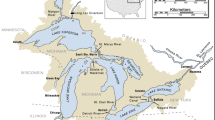
Here we present a 19-year database of physical, chemical, and biological data that span primary producers to secondary consumers measured during summers in 28 lakes in the Adirondack Park in New York State, USA (Fig. 1). The Adirondack Park is a protected state park in northeastern New York that encompasses c. 26,000 km2 of public and private land, and nearly 3000 lakes (> 0.4 ha)23. These lakes are poorly buffered due to surficial and bedrock geology, making them highly susceptible to acidification24,25. Due to the proximity to industrial centers in the mid-western US and prevailing winds26, the region received elevated atmospheric sulfur and nitrogen deposition, which has decreased in recent years27. This unique combination of geology and geography of the Adirondacks resulted in widespread and severe acidification of surface waters, which are now undergoing recovery. Concurrently, the northeastern U.S. has experienced substantial increases in temperature and precipitation and extreme events associated with changing climate28. These stressors individually may have contrasting impacts on aquatic ecosystems. For example, warming surface temperatures and increased thermal stability are predicted to decrease zooplankton species richness29, while recovery from acidification is associated with increases in zooplankton richness30,31.
The dataset presented here is a long-term, comprehensive record of physical, chemical, and biological measurements of a diverse set of lakes undergoing the effects of a changing climate while recovering from acidification. It is a harmonization of multiple open and unpublished data sources, including the Adirondack Effects Assessment Program (AEAP) Aquatic Biota Survey (www.rpi.edu/dept/DFWI/research/aeap/aeap_research.html), the Adirondack Long Term Monitoring Program (ALTM; www.adirondacklakessurvey.org), and the North American Land Data Assimilation System (NLDAS; http://ldas.gsfc.nasa.gov/nldas), and represents a more diverse, long-term data record of Adirondack lakes than has been previously available.
Methods
Site description
The 28 lakes in this dataset are located in the southwestern portion of the Adirondack Park in New York, USA (Fig. 1). This area received the highest rates of atmospheric deposition in the Adirondack Mountains32. When combined with inherently low acid neutralizing capacity (ANC)24,25, high rates of acidic deposition resulted in severe acidification of surface waters in this region33,34. The study lakes are located in five of the six major sub-drainage basins in the Adirondack region and span a range of size, depth, watershed area and hydrologic type (Table 1). The hydrologic classification scheme used was developed by (ref. 35) and is based upon a combination of hydrology (drainage or mounded seepage lakes), underlying geology (thickness of glacial till, or presence of calcite in the basin), and dissolved organic carbon (DOC) concentration (high or low), which combined characterize sensitivity to acidification of each lake. Of the 28 lakes, 20 are thin-till, drainage lakes, the class considered the most sensitive to acidification. Of these 20 thin-till drainage lakes, two have historically high DOC concentrations (TDH), while the remaining 18 have historically low DOC concentrations (TDL). There are six medium-till drainage lakes, two with historically high DOC concentrations (MDH) and four with historically low DOC concentrations (MDL). There is a single mounded seepage lake with historically low DOC (MSL) and one lake drains a watershed with deposits of carbonate (C), which eliminates sensitivity to acidification due to high ANC.
The lakes in this dataset were included in two independent long-term monitoring programs that were established to assess the effects of acid deposition in Adirondack lakes; the Adirondack Effects Assessment Program Aquatic Biota Study (hereafter referred to as AEAP) and the Adirondack Long Term Monitoring Program (hereafter referred to as ALTM). While both programs sampled more lakes than the 28 included in this dataset, these 28 lakes represent the overlap between the two separate programs and thus provide a comprehensive view of the long-term physical, chemical and biological characteristics of each lake. The data record starts in 1994 for all lakes and ends in 2006 for half of the lakes and in 2012 for the remaining half (Table 1). The physical, nutrient and biological data presented here were collected and analyzed by the AEAP. Additional water chemistry data were collected and analyzed as part of the on-going ALTM program. Because these monitoring programs were independent there is overlap in the measured water chemistry analytes. For analytes that were measured by both programs, we selected the data from a single program based upon completeness of record. Overlapping water chemistry measurements (i.e., those not selected from inclusion) can be found in the original data files (Data Citation 1; ‘data_inputs’) but not in the harmonized, final dataset presented here.
Field collection methods
Sampling schedule (AEAP and ALTM)
As part of the AEAP, lakes were sampled three times during the summer (July, Aug, September) from 1994–1996. Starting in 1997, lakes were sampled twice per year (July and August). The ALTM program collected water chemistry data monthly, 12 months of the year starting in 1992 and is an on-going monitoring program (http://www.adirondacklakessurvey.org/). For the purposes of this paper the ALTM monthly chemistry data range from January 1994 to December 2012. For clarity of data sources we note the original program (AEAP or ALTM) that each data type in the subheadings below.
Physical characteristics (AEAP)
Temperature, dissolved oxygen (DO) and photosynthetically active radiation (PAR) measurements were taken at 1 m intervals throughout the entire water column in the deepest spot in each lake as part of the AEAP program. Temperature and DO were measured with a YSI Model 54 meter using a calibrated membrane electrode and thermistor (YSI, Yellow Springs, OH, USA). The thermocline depth was determined in the field as the depth at which the water temperature decreased≥2 °C in a meter. The thermocline depth determined the depths of epilimnetic samples for other variables (e.g., phytoplankton abundance and taxonomy). Secchi disk depth was also measured on each sampling occasion.
Chlorophyll and nutrient concentrations (AEAP)
Water samples to measure chlorophyll a, total nitrogen (TN), total phosphorus (TP), total filterable phosphorus (TFP), and molybdate reactive phosphorus (MRP) were collected at each study site coincident with the collection of the physical data (described above) and biological samples (see below). For sampling occasions when the water column was thermally stratified (as determined by the temperature profile) an integrated epilimnetic sample was collected with a 2.54-cm diameter hose. For un-stratified sampling events, a single integrated sample from the surface to 1 m above the bottom was collected. Samples were stored in high-density amber polyethylene bottles and transported in chilled coolers to the Keck Laboratory at Rensselaer Polytechnic Institute Troy, NY for processing and analysis36,37.
Water chemistry (ALTM)
The ALTM collected water samples for a suite of water chemistry parameters (Table 2). Samples were collected in two different ways depending on the mode of physical access and hydrology of the lake. For all lakes that were accessed by a helicopter and any lake without a surface outlet (see Table 1), samples were collected near the deepest part of the lake at 0.5 m below the surface with a Kemmerer sampler. For all other sites, water samples were collected at the lake outlet to allow safe sampling during periods of thin ice cover and because of limited helicopter availability. Samples were collected in high-density polyethylene bottles and transported in chilled coolers to the Adirondack Lakes Survey Corp. laboratory in Ray Brook, NY for processing and analysis38.
Phytoplankton (AEAP)
A single phytoplankton sample was collected in the deepest part of each lake from the surface down to the 1% PAR (estimated at twice the Secchi depth) with a 2.54-cm diameter integrated hose. For lakes shallower than the estimated 1% PAR depth, samples were collected from the surface to 1 m above the bottom. This approach contrasts with the methodology used for nutrients and chlorophyll a, which were collected as an integrated sample in the epilimnion. A 250-ml subsample of an integrated sample was preserved in the field with a 3% mixture of equal parts glutaraldehyde and formaldehyde for later enumeration and identification of species.
Zooplankton (AEAP)
Replicate zooplankton samples were collected in the deepest part of each lake from surface to 1 m above the bottom or to the depth where DO was<2 mg/L, whichever was shallower, using a hose-integration technique and constant-flow pump. The hose was lowered through the water column at a constant rate and at least 100 L were pumped from each lake (150–200 L for lakes identified as having low zooplankton densities) and concentrated with a 64- μm mesh. Zooplankton were narcotized with carbonated water and immediately preserved in the field with buffered formaldehyde.
Sample Processing
Water chemistry (ALTM)
Aliquots of water samples were divided as necessary for the measurement of each analyte following standard methods outlined in Table 2 and briefly described here. Water color was determined on an unfiltered water sample by visual comparison to a platinum-cobalt standard. Conductivity, pH and ANC were measured electrometrically using a calibrated Orion or YSI glass electrode. Conductivity and pH where measured directly, with pH measured in the field immediately after collection, while ANC was measured using Gran titration39.
A Technicon Autoanalyzer (Seal Analytical, Inc., Mequon, Wisconsin, USA) was used for colorimetric determination of concentrations of ammonium (NH4+), reactive silica (SiO2), total monomeric aluminum (AlTM) and organic monomeric aluminum (AlOM) on an aliquot; the NH4+ samples were acidified with sulfuric acid prior to colorimetric analysis. Inorganic monomeric aluminum (AlIM) concentration was estimated as the difference between AlTM and AlOM (refs 40,41). Major anions including sulfate (SO42−), nitrate (NO3−), fluoride (F−) and chloride (Cl−) concentrations were measured chromatographically42,43 with a Dionex ICS-1100 ion chromatograph (Thermo Fischer Scientific, Waltham, MA, USA). Base cations including sodium (Na+), potassium (K+), magnesium (Mg2+), and calcium (Ca2+) were measured with a PinAAcle 900H atomic absorption spectrophotometer (PerkinElmer, Waltham, MA, USA)44. Total dissolved aluminum (AlTD) was also measured with a PinAAcle 900H atomic absorption spectrophotometer but one fitted with a high-temperature graphite furnace and an AS900 auto sampler (PerkinElmer, Waltham, MA) to volatilize the inorganic and organic Al complexes40,45. A Tekmar Dorhmann Pheonix 8000 carbon analyzer (Teledyne Tekmar, Mason, OH, USA) was used to measure concentrations of dissolved organic and inorganic carbon (DOC and DIC, respectively) by converting the carbon in the sample to carbon dioxide and measuring the carbon dioxide with an infrared spectroscopic sensor45. A filtered aliquot (0.45 micron pore size GFF), preserved with phosphoric acid, was used to determine DOC via UV persulfate oxidation. DIC was measured in a separate sealed water sample collected in the field to ensure the DIC was not lost to the atmosphere and therefore underestimated46.
Chlorophyll and nutrient concentrations (AEAP)
As with the water chemistry, aliquots of water samples were divided as necessary and measured with standard methods outlined in Table 2 and described here. Chlorophyll a concentration was determined by filtering water sampled onto a glass fiber filter, extracting the chlorophyll a in 90% acetone for 4–24 h and measuring fluorescence with a Turner MODEL 10- AU fluorometer47 (Turner Designs, Sunnyvale, CA, USA).
Total nitrogen (TN) and total phosphorus (TP) concentrations were measured on a well-mixed unfiltered aliquot of lake water while total filterable phosphorus (TFP) was measured on filtrate passed through a 0.45-micron membrane filter. TN was measured using persulfate oxidation48. For TP and total filterable phosphorus (TFP) concentrations, aliquots were digested in a potassium persulfate solution via autoclave at high heat, then determined colorimetrically using a spectrophotometer45. Molybdate reactive P (MRP) and ammonium (NH4+) were measured on raw water samples. While this differs slightly from the standard methods, particulates are so low in these lakes that using unfiltered samples should have had little effect on the outcome. Both MRP and NH4+ were measured colorimetrically via flow injection (Lachat QuikChem Flow Injection Analysis System, Hach Company, Loveland, CO, USA)45. Note that NH4+ appears in both the nutrient and water chemistry data sets. The same procedure was used to estimate NH4+ concentration but the location and depth of the samples differed. The AEAP data set measure NH4+ concentration from an integrated epilimnetic sample near the deep spot while the ALTM measured NH4+ concentration at 0.5 m near the deep spot or at the lake outlet depending upon the lake (see Table 1 for details).
Phytoplankton (AEAP)
Phytoplankton samples from 1994 and 1995 were analyzed at the University of Louisville (Louisville, Kentucky, USA). All samples from 1996 or later were analyzed at the Patrick Center for Environmental Research at the Academy of Natural Sciences of Drexel University (Philadelphia, Pennsylvania, USA) hereafter referred to as ANS. At the University of Louisville, samples were filtered onto a membrane filter, cleared and mounted under a coverslip on a microscope slide49. One to three slides were prepared for each sample and 10–30 fields per slide were examined under 625x magnification. At ANS the samples were concentrated by centrifuge and examined under 538x magnification with an inverted microscope using Utermöhl sedimentation technique and counting random fields50,51. Approximately 500 natural units were enumerated for each sample. Identifications of phytoplankton were made to the species level when possible using keys52–59. All taxonomy was updated according to60 as of October 2017. All taxonomic information and updates are shown in the phytoplankton reformat table (Data Citation 1, ‘data_inputs’ folder).
To determine biovolumes of algal taxa a simple geometric shape was matched to an individual cell, 1 to 3 dimensions of the cell were measured and these measurements were used to calculate the volume (in μm3). Fifteen specimens were measured for each taxon with additional measurements for larger and variably sized taxa. In several cases of rare taxa, fewer specimens were measured and/or sizes were determined from literature values.
Zooplankton (AEAP)
Crustacean zooplankton were counted in a Bogorov chamber under 60x magnification using standard subsampling techniques with subsamples 1–5 ml in volume61,62. Rotifers were counted in a Sedgewick-Rafter cell (1 ml subsample) under 100x magnification. All individuals were identified to species when possible. Crustacean zooplankton were identified using keys from (refs 63–66), while rotifers were identified using66–70. All zooplankton counts and identifications were identified and counted under the supervision of W.H. Shaw with the exception of 2000–2002 when samples were counted at Marist College (Poughkeepsie, New York, USA) using standard methods. Taxonomy of the original dataset has been revised to reflect updated classification as of January 2017 based on (refs 71–73). Taxonomic updates and information are in separate ‘reformat tables’ for crustacean zooplankton and rotifers for ease of updating in the future (Data Citation 1, ‘data_inputs’ folder).
Zooplankton biomass was estimated from the count data using published empirical length-weight relationships (crustacean zooplankton) or formulas for body volume calculations (rotifers) for the freshwater zooplankton species in the dataset or for congeners when necessary. For the rotifers, body volume formulas are from (refs 74,75). Length-weight regressions for the crustacean zooplankton are from (refs 75–81).
Since size measurements were not taken of the zooplankton during the enumeration procedure, we used average organism lengths for each species from published studies or from the North Temperate Lakes Long-Term Ecological Research site (NTL LTER; https://lter.limnology.wisc.edu/data). The values derived from the NTL LTER are the average length of all individuals within a species collected across all seven lakes from 1982-2015 (year-round sampling). Measurements for an additional seven crustacean species are from (refs 76,77,82–84), or from a small glacial lake in the Poconos Mountains of Northeastern PA (T. Leach unpublished data). Measurements for rotifers are from (refs 74,75) or the North Temperate Lakes LTER zooplankton dataset 1982–2015 (http://lter.limnology.wisc.edu). See Data Citation 1, zoop_biomass_conversion.csv for all equations, measurements, and references.
Published length-weight relationships for the crustacean zooplankton typically incorporate dry weights of an individual. For consistency with rotifers and phytoplankton biomass estimates, we converted dry weights to wet weights by assuming that the ratio of dry:wet weight was 0.1(10%) following (ref. 85). For the phytoplankton and some of the rotifer species, the estimates are expressed as volume (mm3 individual−1). For comparison with the crustacean zooplankton biomass, biovolume was converted to biomass by assuming that all organisms had a density of 1 (ref. 85); from this assumption organism volume as μm3 individual−1 is equivalent to biomass as μg individual−1.
Meteorological data
Long-term meteorological data, including air temperature, relative humidity, wind speed, and downwelling shortwave and longwave radiation, were extracted from the North American Land Data Assimilation System (NLDAS) from 1979–2012 using the geographic location of each lake. NLDAS is a gridded reanalysis of historic weather data over North America produced and maintained as a collaboration between NASA and NOAA (http://ldas.gsfc.nasa.gov/nldas). Meteorological data from NLDAS were averaged (simple mean) to represent daily values for each variable.
Data harmonization
We harmonized the different data sources using a combination of lake names and latitude/ longitude records. We verified all lake names against the Geographical Names Information System database (https://nhd.usgs.gov/gnis.html) using latitude and longitude reference. Further, to connect the dataset with a physical water body, we linked each site with its corresponding polygon in the high-resolution U.S. Geological Survey’s National Hydrography Dataset (NHD) and include corresponding polygons and permanent identifiers for future use. Sampling date formats and lake names were also standardized so that data files can be easily linked by lake and sampling occasion in addition to permanent identifiers. See Fig. 2 for a detailed workflow and relationship between each data type.
Input files on the far left, R code scripts in the grey boxes and output files (.csv format) are on the right. Information on the output files can be found in Table 3. All original data files and scripts to re-create each file are available at Data Citation 1.
Code availability
All key harmonization and data conversion steps were done in the R scientific computing language version 3.3.3 (ref. 86). For reference, original data files and all harmonization R code are included in a Data Citation 1, ‘data_input’ and ‘Code_toclean’ folders, respectively.
Data Records
The data are available in two formats; as comma separated files (.csv) within the folder ‘data’ (Data Citation 1) and as an R Data Package wrapper, adklakedata (Data Citation 2), which automatically retrieves and makes the data files available in the R programming environment86. Both the ‘data’ folder within Data Citation 1 and the adklakedata package contain the same data.
There are several different categories of data in the dataset: (1) geographic, (2) physical, (3) water chemistry, (4) biological, (5) meteorological and (6) other (Table 3, Fig. 2). Additionally, each.csv data file has an accompanying text file with the same name that contains a description of each column header, units of each variable and other pertinent metadata. Data are split across files containing different types of data based on data structure but all data files contain a column with the unique lake name and date on which the data were measured, which enables linking data files together for analysis (See data Usage from more information). A list with a description of the files associated with the dataset is provided in ‘adklake_data_descriptions.txt’ and Table 3. This information is also available in the adklakedata documentation available on CRAN, the Comprehensive R Archive Network (https://cran.r-project.org/).
Technical Validation
There were two types of technical validation performed on these data. The first involved extensive quality assessment and quality control (QA/QC) of the data collection and sample analysis methods. The second included validation of the data cleaning and harmonization to create a unified and compatible data structure across all data types.
Data collection and sample processing validation
A QA/QC program for the AEAP chlorophyll a and nutrient samples consisted of running a certified external standards every 10th sample, spikes in 10% of samples to verify analyte recovery and replication of sample analysis for 10% of samples. Standard curves were run at the beginning and end of every analyte analysis batch (20 samples) as well as a blank and standard in the middle of each run to assess drift and develop the standard curve. If the value of these standards was not within 10% of expected the entire batch was re-analyzed. The Keck Laboratory where the AEAP samples were analyzed was Environmental Laboratory Accreditation Program and National Environmental Laboratory Accreditation Conference certified and participated in the USGS Standard Water Sample Program administered by the Environment Canada, National Water Research Institute Ecosystem Inter-laboratory Quality Assurance Program. As part of this program, proficiency samples were analyzed every six months to assure quality control and the laboratory was audited every two years.
QA/QC procedures for all samples counted at the ANS included recounts of specific samples and consultation with outside experts for verification of taxonomy. Outside experts included Dr Ann St. Amand (PhytoTech, St. Joseph, Michigan, USA), Dr Rex. L. Lowe (Bowling Green State University, Bowling Green, Ohio, USA) and William R. Cody (Aquatic Taxonomy Specialists, Malinta, Ohio, USA). Dr Ann St. Amand also verified taxonomy of samples counted at the University of Louisville. Also, images were taken of most taxa to help insure consistency of identifications from the beginning to the end of the project, especially for undescribed taxa.
To ensure consistent species identification of the zooplankton, photographs were taken for both rotifers and crustaceans. When possible, microscope slides were created for crustaceans showing important anatomic criteria. Calanoid and cyclopoid species identifications were verified with prepared slide mounts of antenna and 5th leg preparations of both males and females when possible. When congeneric species or multiple species of Daphnia were present, slides of 25 randomly selected individuals were prepared to estimate the relative proportions of each congener and compared to proportions within full counts. Identification of reoccurring but rare rotifer species were verified by Dr Richard Stemberger (Dartmouth College, Hanover, NH). For all zooplankton samples the subsample-to-sample ratio was maximized in order to limit multiplication errors and improve accuracy of counts. Duplicates counts were performed on samples from every tenth lake during the 1994–1996 and 2001–2002 sampling periods. These duplicate counts showed that counting precision was high.
The water chemistry data collected as part of the ALTM sampling program also had a QA/QC program in place to assure data quality and measurement accuracy. This procedure included a clear line of sample custody, standard maximum holding times and assessment of analytical precision. To assess analytical precision, 5% of all samples were collected and analyzed in triplicate. On days when field triplicates were collected the values in the dataset represent the average of those triplicates. Laboratory duplicates (i.e., samples split in the laboratory from the same field collection container) were analyzed every 20th sample. Field blanks were also created for at least 5% of the total field samples. Field blanks were prepared in the laboratory by filling collection containers with deionized water and then processing them in the field as though they were field samples. Analytical standards were run at the beginning of a batch and every tenth sample. Only field samples bracketed by passing standards were accepted. Correction actions such as recalibration and sample re-runs were performed if coefficient of variation between laboratory duplicates or standards was greater than 0.1.
Data harmonization validation
The R code to restructure, and harmonize the data, as well as the original data files are included in the folders called ‘Code_toclean’ and ‘input_data’ in Data Citation 1. All of the code was written by T. Leach and reviewed by L. Winslow. A series of manual QA/QC steps were performed to verify that there were no data processing errors between the raw source files and final data tables. A random 1% of each data type was manually checked between the original and final data files. All physical data including temperature and dissolved oxygen profiles and Secchi disk depths were manually checked for out of range or unexpected values. Out of range values were corrected or removed where appropriate. The database and R code were revised as needed throughout these manual validation steps to correct mistakes.
Usage Notes
The combined dataset is distributed as a series of comma separated value (CSV) files that contain the data organized by data type (See Table 3 for description of each data type). Despite being separate files, all data can be linked by geographic location (site) using ‘lake.name’ or ‘PERMANENT_ID’ (from the NHD), or on a temporal axis using the ‘date’ variable. Keep in mind that not all chemical, physical and/or biological data were collected on the same day so a matching window (for example±7 days) may be useful to employ when merging different data types for analysis.
We have developed two methods for data access. One, the CSV files of all data can be downloaded directly from an online repository (Data Citation 1). This supports general use cases, as CSV is a common and widely supported data format. Two, we have developed an R package wrapper for the dataset that is available from CRAN, the Comprehensive R Archive Network. This package adklakedata automates the downloading, local storage, and access of the data. Data are accessed using the ‘adk_data’ function which accepts a parameter for each dataset (e.g., `adk_data(‘tempdo’)’ for temp and dissolved oxygen data).
Additional information
How to cite this article: Leach, T. H. et al. Long-term dataset on aquatic responses to concurrent climate change and recovery from acidification. Sci. Data 5:180059 doi: 10.1038/sdata.2018.59 (2018).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
References
O’Reilly, C. M. et al. Rapid and highly variable warming of lake surface waters around the globe. Geophys. Res. Lett. 42, 10773–10, 781 (2015).
Winslow, L. A., Read, J. S., Hansen, G. J. A. & Hanson, P. C. Small lakes show muted climate change signal in deepwater temperatures. Geophys. Res. Lett. 42, 355–361 (2015).
Likens, G. E., Bormann, F. H. & Johnson, N. Acid rain. Environment 14, 33–40 (1972).
Strock, K. E., Nelson, S. J., Kahl, J. S., Saros, J. E. & McDowell, W. H. Decadal trends reveal recent acceleration in the rate of recovery from acidification in the northeastern U.S. Environ. Sci. Technol. 48, 4681–4689 (2014).
Driscoll, C. T., Driscoll, K. M., Fakhraei, H. & Civerolo, K. Long-term temporal trends and spatial patterns in the acid-base chemistry of lakes in the Adirondack region of New York in response to decreases in acidic deposition. Atmos. Environ. 146, 5–14 (2016).
Monteith, D. T. et al. Dissolved organic carbon trends resulting from changes in atmospheric deposition chemistry. Nature 450, 537–540 (2007).
Smith, V. H. et al. Comment: Cultural eutrophication of natural lakes in the United States is real and widespread. Limnol. Oceanogr. 59, 2217–2225 (2014).
Bennett, E. M., Carpenter, S. R. & Caraco, N. F. Human Impact on Erodable Phosphorus and Eutrophication: A Global Perspective. Bioscience 51, 227–234 (2001).
Simberloff, D., Parker, I. M. & Windle, P. N. Introduced species policy, management, and future research needs. Front. Ecol. Environ. 3, 12–20 (2005).
Shaker, R. R. et al. Predicting aquatic invasion in Adirondack lakes: A spatial analysis of lake and landscape characteristics. Ecosphere 8, e01723 (2017).
Zohary, T. & Ostrovsky, I. Ecological impacts of excessive water level fluctuations in stratified freshwater lakes. Inl. Waters 1, 47–59 (2011).
Likens, G. E. & Bormann, F. H. Acid Rain: A Serious Regional Environmental Problem. Science 184, 1176–1179 (1974).
Stoddard, J. L. et al. Regional trends in aquatic recovery from acidification in North America and Europe. Nature 401, 575–578 (1999).
Michelena, T. M. et al. Aluminum toxicity risk reduction as a result of reduced acid deposition in Adirondack lakes and ponds. Environ. Monit. Assess. 188, 636 (2016).
Foley, B., Jones, I. D., Maberly, S. C. & Rippey, B. Long-term changes in oxygen depletion in a small temperate lake: Effects of climate change and eutrophication. Freshw. Biol 57, 278–289 (2012).
Bertram, P. E. Total Phosphorus and Dissolved Oxygen Trends in the Central Basin of Lake Erie, 1970–1991. J. Great Lakes Res. 19, 224–236 (1993).
Heisler, J. et al. Eutrophication and harmful algal blooms: A scientific consensus. Harmful Algae 8, 3–13 (2008).
Editorial. Long-term relationships. Nat. Ecol. Evol 1, 1209–1210 (2017).
Winslow, L. A., Read, J. S., Hansen, G. J. A., Rose, K. C. & Robertson, D. M. Seasonality of change: Summer warming rates do not fully represent effects of climate change on lake temperatures. Limnol. Oceanogr. 62, 2168–2178 (2017).
Lottig, N. R. et al. Long-Term Citizen-Collected Data Reveal Geographical Patterns and Temporal Trends in Lake Water Clarity. PLoS ONE 9, 155–165 (2014).
Woolway, R. I. et al. Warming of Central European lakes and their response to the 1980 s climate regime shift. Clim. Change 142, 505–520 (2017).
Kritzberg, E. S. Centennial-long trends of lake browning show major effect of afforestation. Limnol. Oceanogr. Lett 2, 105–112 (2017).
Colquhoun, J., Kretser, W. & Pfeiffer, M. Acidity Status Update of Lakes and Streams in New York State. (Report No. WM P-83 (New York State Department of Environmental Conservation, 1984).
Omernik, J. & Powers, C. Total alkalinity of surface waters: a national map. Ann. Assoc. Am. Geogr 73, 133–136 (1983).
Jenkins, J. & Keal, A. . The Adirondack atlas: a geographic portrait of the Adirondack Park (Syracuse University Press, 2004).
Driscoll, C. T. et al. Acidic deposition in the northeastern United States: Sources and inputs, ecosystem effects, and management strategies. Bioscience 51, 180–198 (2001).
Galloway, J. N., Likens, G. E. & Hawley, M. E. Acid Precipitation: Natural Versus Anthropogenic Components. Science 226, 829–831 (1984).
Melillo, J. M., Richmond, T. & Yohe, G. W. Climate Change Impacts in the United States Climate Change Impacts in the United States: The Third National Climate Assessment. doi:10.7930/j0z31WJ2 (2014).
Shurin, J. B. et al. Environmental stability and lake zooplankton diversity - contrasting effects of chemical and thermal variability. Ecol. Lett. 13, 453–463 (2010).
Keller, W. & Yan, N. D. Recovery of Crustacean Zooplankton Species Richness in Sudbury Area Lakes following Water Quality Improvements. Can. J. Fish. Aquat. Sci. 48, 1635–1644 (1991).
Keller, W., Yan, N. D., Somers, K. M. & Heneberry, J. H. Crustacean zooplankton communities in lakes recovering from acidification. Can. J. Fish. Aquat. Sci. 59, 726–735 (2002).
Jenkins, J., Roy, K., Driscoll, C. T. & Buerkett, C. Acid Rain and the Adirondacks: A Research Summary (Adirondack Lake Survey Corporation, 2005).
Fakhraei, H. et al. Development of a total maximum daily load (TMDL) for acid-impaired lakes in the Adirondack region of New York. Atmos. Environ. 95, 277–287 (2014).
Driscoll, C. T., Newton, R. M., Gubala, C. P., Baker, J. P., Christensen, S. W. Adirondack Mountainsin Acidic Deposition and Aquatic Ecosystems (ed. Charles D. F. ) (Springer, 1991).
Driscoll, C. T. & Newton, R. M. Chemical characteristics of Adirondack lakes. Environ. Sci. Technol. 19, 1018–1024 (1985).
Nierzwicki-Bauer, S. et al. Adirondack Effects Assessment Program Final Report: summary of aquatic biota and watershed integrated nitrogen cycling studies (2006).
Nierzwicki-Bauer, S. A. et al. Acidification in the adirondacks: Defining the biota in trophic levels of 30 chemically diverse acid-impacted lakes. Environ. Sci. Technol. 44, 5721–5727 (2010).
Adirondack Lake Survey Corp. Standard Operating Procedures Laboratory Operations (Adirondack Lake Survey Corp., 2002).
USEPA. Handbook of Methods for Acidic Deposition Studies: Laboratory Analysis for Surface Water Chemistry. USEPA Report No. EPA-600-4-87-026 (US Environmenal Protection Agency, 1987).
Driscoll, C. T. A Procedure for the Fractionation of Aqueous Aluminum in Dilute Acidic Waters. Int. J. Environ. Anal. Chem. 16, 267–283 (1984).
McAvoy, D. C., Santore, R. C., Shosa, J. D. & Driscoll, C. T. Comparison between Pyrocatechol Violet and 8-Hydroxyquinoline Procedures for Determining Aluminum Fractions. Soil Sci. Soc. Am 56, 449–455 (1992).
Small, H., Stevens, T. S. & Bauman, W. C. Novel Ion Exchange Chromatographic Method Using Conductimetric Detection. Anal. Chem. 47, 1801–1809 (1975).
USEPA. Methods for the Determination of Inorganic Substances in Environmental Samples. USEPA Report No. EPA-600/R-93/100 (US Environmental Protection Agency, 1993).
Slavin, W. Atomic Absorption Spectroscopy (Wiley-Interscience, 1968).
Kopp, J. F. & McKee, G. D. Methods for chemical analysis of water and wastes. USEPA Report No. EPA-600/4-79-020 (US Environmental Proteciton Agency, 1983).
Dohrmann. Automated Laboratory Total Organic Carbon Analyzer Operatoral Manual. (1984).
Turner, G. K. Fluorometryin Bioluminescence and Chemiluminescence Instruments and Applications: Vol. 1 (ed. Van Dyke K. 241 (CRC Press, Inc., 1985).
Langner, C. L. & Hendrix, P. F. Evaluation of a persulfate digestion method for particulate nitrogen and phosphorus. Water Res 16, 1451–1454 (1982).
Bukaveckas, P. A. Effects of calcite treament on primary producers in acidified Adirondack Lakes. II. Short-term response by phytoplankton communities. Can. J. Fish. Aquat. Sci. 46, 352–359 (1989).
Charles, D. F., Knowles, C. A. & Davis, R. S. Protocols for the analysis of algal samples collected as part of the U.S. Geological Survey National Water-Quality Assessment Program. Report No. 02-06 (Academcy of Natural Sciences. 2002).
Hasle, G. R. in Phytoplankton Manual (United Nations Educational, Scientific and Cultural Organization, 1978).
Prescott, G. W. Algae of the Western Great Lakes (WM.C. Brown Company Publishers, 1962).
Campbell, P. H. Studies of brackish water phytoplankton (UNC Sea Grant Program, 1973).
Croasdale, H. Freshwater algae of Ellsmere Island, N. w. t. (exclusive of diatoms and flagellates). Publ. Bot 1973, 131 (1973).
Whitford, L. A. & Schumacher, G. J. A Manual of Fresh-Water Algae (Sparks Press, 1973).
Croasdale, H. T., Bicudo, C.E. de M & Prescott, G. W. Synopsis of North American Desmids: Part II Desidiaceae-Placodermae Section 5 (University of Nebraska Press, 1983).
Dillard, G. E. Freshwater algae of the Southeastern United States (J. Cramer, 2000).
Prescott, G. W., Croasdale, H. T., Vinyard, G. L. & Bicudo, C. E. de M . Synopsis of North American Desmids: Part II Desidiaceae - Placodermae Section 4 (University of Nebraska Press, 1982).
Uherkovich, G. Die Scenedesmus-Arten Ungarns (Akadémiai Kiadó, 1966).
Guiry, M. D. & Guiry, G. M. AlgaeBasehttp://www.algaebase.org (2017).
Gannon, J. E. Two Counting Cells for the Enumeration of Zooplankton Micro-Crustacea. Trans. Am. Microsc. Soc 90, 486–490 (1971).
Bukaveckas, P. A. & Shaw, W. Effects of base addition and brook trout (Salvelinus fontinalis) introduction on the plankton community of an acidic Adirondack lake. Can. J. Fish. Aquat. Sci. 54, 1367–1376 (1997).
Blacer, M. D., Korda, N. L. & Dodson, S. I. Zooplankton of the Great Lakes: A Guide to the Identification and Ecology of the Common Crustacean Species (University of Wisconsin Press, 1984).
Herbert, P. D. N. The Daphnia of North America: An illustrated fauna (University of Guelph, 1995).
Haney, J. An Image-based Key to the Zooplankton of the North Americahttp://cfb.unh.edu/cfbkey/html/index.html (2017).
Edmondson, W. T. Fresh-water Biology (John Wiley & Sons, Inc., 1959).
Pennak, R. W. Freshwater Invertebrates of the United States (Ronald Press Co., 1953).
Ruttner-Kolisko, A. Das zooplankton der Benningewasser: III Rototoria. Die Bennengewasser 26, 99–234 (1972).
Pontin, R. A. A key to British Freshwater Planktonic Rotifera (Freshwater Biological Association, 1978).
Stemberger, R. S. A Guide to Rotifers of the Laurentian Great Lakes. USEPA Report No. EPA 600/4 79-021 (US Environmental Protection Agency, 1979).
Richter, S., Braband, A., Aladin, N. & Scholtz, G. The phylogenetic relationships of ‘predatory water-fleas’ (Cladocera: Onychopoda, Haplopoda) inferred from 12 S rDNA. Mol. Phylogenet. Evol. 19, 105–113 (2001).
Jersabek, C. D. & Leitner, M. F. The Rotifer World Catalog. http://rotifera.hausdernatur.at/ (2013).
Board, W. E. World Reister of Marine Species. VLIZ. 10.14284/170 (2017).
Ejsmont-Karabin, J. Empirical equations for biomass calcualtions of planktonic rotifers. Pol. Arch. Hydrobiol. 45, 513–522 (1998).
Bottrell, H. H. et al. A review of some problems in zooplankton production studies. Nor. J. Zool. 24, 419–456 (1976).
Dumont, H. J., Velde, I. & Dumont, S. The dry weight estimate of biomass in a selection of Cladocera, Copepoda and Rotifera from the plankton, periphyton and benthos of continental waters. Oecologia 19, 75–97 (1975).
Culver, D. A., Boucherle, M. M., Bean, D. J. & Fletcher, J. W. Biomass of Freshwater Crustacean Zooplankton from Length–Weight Regressions. Can. J. Fish. Aquat. Sci. 42, 1380–1390 (1985).
Persson, G. & Ekbohm, G. Estimation of dry weight in zooplankton populations: methods applied to crustacean populations from lakes in the Kuokkel Area, Northern Sweden. Arch. Fur Hydrobiol 89, 225–246 (1980).
O’Brien, W. J. & DeNoyelles, F. J. Relationship between nutrient concentration, phytoplankton density and zooplankton density in nutrient enriched experimental ponds. Hydrobiologia 44, 105–125 (1974).
Rosen, R. A. Length-Dry Weight Relationships of Some Freshwater Zooplanktona. J. Freshw. Ecol. 1, 225–229 (1981).
Watkins, J., Rudstam, L. & Holeck, K. Length-weight regressions for zooplankton biomass calculations – A review and a suggestion for standard equations (Cornell Biological Field Station, 2011).
Wicklum, D. Variation in horizontal zooplankton abundance in mountain lakes: shore avoidance or fish predation? J. Plankton Res. 21, 1957–1975 (1999).
Hansen, M. J. & Wahl, D. H. Selection of Small Daphnia pulex by Yellow Perch Fry in Oneida Lake, New York. Trans. Am. Fish. Soc 110, 64–71 (1981).
Dieguez, M. C. & Gilbert, J. J. Suppression of the rotifer Polyarthra remata by the omnivorous copepod Tropocyclops extensus: predation or competition. J. Plankton Res. 24, 359–369 (2002).
Pace, M. L. & Orcutt, J. D. The relative importance of protozoans, rotifers, and crustaceans in a freshwater zooplankton community. Limnol. Oceanogr. 26, 822–830 (1981).
R Developement Core Team. R: A Language and Environment for Statistical Computing. R Found. Stat. Comput. 1, 409 (2015).
Roy, K., Houck, N., Hyde, P., Cantwell, M. & Brown, J. The Adirondack Long-Term Monitoring Lakes: A compendium of site descriptions, recent chemistry and selected research information. NYSERDA Report No. 11-12 (New York State Energy Research and Development Authority, 2011).
Data Citations
Winslow, L. A., & Leach, T. H. figshare https://doi.org/10.6084/m9.figshare.5686987.v2 (2018)
Winslow, L. A., Leach, T. H., & Hahn, T. Zenodo https://doi.org/10.5281/zenodo.1181754 (2018)
Acknowledgements
Long-term data collection is not possible without the help of countless students, technicians and volunteers. But in particular we would like to thank Scott Quinn, Bryan Weatherwax, Robert Bombard, and the New York State Police Aviation Unit without whose support the AEAP sampling effort would not have been possible. The AEAP program was funded by the U.S. Environmental Protection Agency (US EPA; contract #68D20171) from 1994–2006 and by New York State Energy Research and Development Authority (NYSERDA; contract #16298) since 2006. KCR acknowledges support of NYSERDA contract #115876 and National Science Foundation grant EF 1638704 for the development of this manuscript. The ALTM program was supported by NYSERDA, the New York State Department of Environmental Conservation (NYSDEC), the U.S. EPA and the U.S. Geological Survey and conducted by the Adirondack Lakes Survey Corporation (ALSC). Special thanks to Sara Burke, Michael Cantwell, Sue Capone, Pamela Hyde Corey, Korey Devins, James Dukett, Nathan Houck, Matthew Kelting, Monica Schmidt, and Philip Snyder of the ALSC staff. We would also like to thank Tobi Hahn for help with the R package creation. This paper has not been subjected to US EPA or NYSDEC peer and administrative review; therefore, the conclusions and opinions contained herein are solely those of the authors, and should not be construed to reflect the views of either agency.
Author information
Authors and Affiliations
Contributions
T.H.L. cleaned and compiled all data and led the writing of the manuscript. All authors contributed to and provided feedback on the manuscript. L.A.W. lead the R package creation, and verified data cleaning and compilation code. M.W.S. assisted with data harmonization QA/QC. S.N.B. and J.W.S. designed the study AEAP study with assistance from C.W.B., R.A.D. and L.W.E., and J.W.S. coordinated the AEAP field sampling effort with assistance from J.B. C.W.B., P.A.B., L.W.E., R.A.D., J.L.F., C.A.G., T.M.M., S.N.B., W.H.S., J.W.S., and D.A.W. collected and/or analyzed samples as part of the AEAP, and along with D.F.C. contributed to writing the AEAP methods. F.W.A. also verified the most recent taxonomic updates of phytoplankton data and assisted with data cleaning. C.T.D., C.S.F., and K.M.R. contributed to writing of the ALTM methods.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
ISA-Tab metadata
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files made available in this article.
About this article
Cite this article
Leach, T., Winslow, L., Acker, F. et al. Long-term dataset on aquatic responses to concurrent climate change and recovery from acidification. Sci Data 5, 180059 (2018). https://doi.org/10.1038/sdata.2018.59
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/sdata.2018.59
This article is cited by
-
Changes in acidity, DOC, and water clarity of Adirondack lakes over a 30-year span
Aquatic Sciences (2021)