Abstract
Human locomotion is a complex process that requires the integration of central and peripheral nervous signalling. Understanding the brain’s involvement in locomotion is challenging and is traditionally investigated during locomotor imagination or observation. However, stationary imaging methods lack the ability to infer information about the peripheral and central signalling during actual task execution. In this report, we present a dataset containing simultaneously recorded electroencephalography (EEG), lower-limb electromyography (EMG), and full body motion capture recorded from ten able-bodied individuals. The subjects completed an average of twenty trials on an experimental gait course containing level-ground, ramps, and stairs. We recorded 60-channel EEG from the scalp and 4-channel EOG from the face and temples. Surface EMG was recorded from six muscle sites bilaterally on the thigh and shank. The motion capture system consisted of seventeen wireless IMUs, allowing for unconstrained ambulation in the experimental space. In this report, we present the rationale for collecting these data, a detailed explanation of the experimental setup, and a brief validation of the data quality.
Design Type(s) | observation design • replicate design |
Measurement Type(s) | brain activity measurement • musculoskeletal movement • locomotion involved in locomotory behavior |
Technology Type(s) | electroencephalography • electromyography • full-body motion capture system |
Factor Type(s) | replicate • biological sex • age |
Sample Characteristic(s) | Homo sapiens |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
Human bipedal locomotion results from the complex interaction of dynamic networks in the spinal cord with feedback mechanisms originating from sensory inputs through spinal and supraspinal pathways. Fundamental circuitry in the spinal cord are believed to be responsible for generating basic locomotor patterns, while descending pathways from the brain are thought to provide signalling that can initiate, terminate, and guide locomotion1. Traditionally, many studies on locomotion have relied on invasive experiments in the spinal cord and motor cortices of cats1. However, invasive electrophysiological approaches are not feasible to study the involvement of cortical networks in human walking due to the ethical challenges of invasive recordings in human subjects; thus, the role of the spinal cord and brain during human bipedal locomotion has been investigated using non-invasive imaging modalities2–4. Many studies using functional magnetic resonance imaging (fMRI) and magnetoencephalography (MEG)—both stationary methods—have relied on gait imagery or observation2–4 as a source for understanding cognition and gait. While motor imagery and observation may result in the activation of networks involved in actual task execution5–12, stationary studies do not allow for the investigation of networks connecting cortical brain activity and muscular activation involved in the action. Thus, we must rely on mobile systems for simultaneous collection of information from the central and peripheral nervous systems.
Electroencephalography (EEG) is a powerful tool for studying the brain’s involvement in movement execution with a high temporal resolution. On the down side, EEG lacks the spatial resolution of other techniques and suffers from a significantly attenuated signal due to volume conduction13,14. The localization of the EEG signal sources in the brain can be challenging, since the volume conduction model is comprised of multiple layers, including fluid media and anisotropic tissues13,14. Fortunately, there are methods for extracting highly localized information, even with spatial resolution, and the high temporal resolution provides detailed information related to the task. These characteristics make EEG a suitable source for brain imaging during an overground locomotion task.
EEG has been used to investigate cortical involvement during steady-state walking15–26 and multi-tasking27. However, most of these studies have been limited to treadmills, which fix the cadence and speed of locomotion. In this report, we present a highly comprehensive dataset from ten healthy individuals during a multi-terrain walking task. The subjects were instrumented with non-invasive EEG, EMG, and full body motion capture while walking on a custom gait course including level ground, ramps, and stairs. To the best of our knowledge, this dataset is the first recording of continuous overground walking while the subjects were instrumented with EEG. Furthermore, the addition of muscle and kinematic information enhances the richness of this unique dataset. A major concern of EEG is the presence of physiological and non-physiological artifacts during movement. Previous studies have shown that with a careful setup, motion artifacts are negligible at lower walking speeds28.
Previous datasets have been published with EEG data during seated cognitive tasks29,30, during a stimulus-viewing task after transcranial magnetic stimulation (TMS)31, and during perceptual tasks during simultaneous EEG, fMRI, and MEG32. Luciw et al.33 published a comprehensive dataset containing EEG, EMG, kinematic, and kinetic data from twelve health subjects during a grasp and lift. To our knowledge, this is the first publicly available dataset that includes full brain and body imaging (EEG, EMG, kinematics) during overgound multi-terrain locomotion.
We have previously used this dataset to investigate corticomuscular coherence34, cortical activations during multi-terrain walking35,36, and the neural dynamics associated with transitions between walking terrains37. Additionally, we have used this dataset to demonstrate the ability to continuously predict muscle activation patterns directly from EEG34,38 and joint angles from EMG signals39. The multi-modal nature of this data sets enables future studies in brain-computer interfaces (BCIs) and motor control during human locomotion, such as:
-
Investigation of corticomuscular coherence in the source domain
-
Analysis of motion artifacts in EEG during multi-terrain walking
-
Design of an offline decoder for continuous prediction of joint angles from EEG
-
Biomechanical analysis of gait and muscle activation patterns during multi-terrain walking (e.g., symmetry, stride length)
Methods
Participants
Ten healthy individuals (five males, five females; ages 18-31 years old) with no history of neurological disorder, lower-limb pathology, or gait abnormalities participated in this study. The experimental protocol and informed consent (reviewed and signed by each participant) were approved by the Institutional Review Board (IRB) at the University of Houston. All experiments were performed in accordance with the 45 Code of Federal Regulations (CFR) part 46 (“The Common Rule”), specifically addressing the protection of human study subjects as promulgated by the U.S. Department of Health and Human Services (DHHS).
Instrumentation & Data Collection
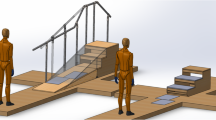
The participants were instrumented with a non-invasive EEG cap, EMG, and full body motion capture for comprehensive brain-body imaging during an over ground multi-terrain locomotion task. Figure 1 shows a fully instrumented subject.
(a) Able-bodied subject instrumented with EEG, EMG, and full-body motion capture. (b) EEG channel montage and EOG locations. EOG_HL: horizontal left, EOG_HR: horizontal right, EOG_VA: vertical above, EOG_VB: vertical below. (c) Close-up image of active EEG electrodes. (d) Experimental gait course including level walking, ramps, and stairs.
Each subject’s head circumference was measured to allow for selection of an appropriately sized EEG. A 64-channel Ag/AgCl active electrode EEG setup (BrainAmp DC and MOVE, Brain Products GmbH, Germany) was used to wirelessly record from the face and scalp (sampling frequency=1000 Hz). The data were recorded using the BrainVision Recorder software (Brain Products GmbH, Germany). Channels TP9, PO9, PO10, and TP10 were removed from the cap and used for electrooculography (EOG) to capture blinks and eye movements. The remaining 60 channels were arranged according to the modified international 10–20 system with slight modifications. Electrodes at channel locations FT9 and FT10 were moved to locations AFz and FCz, respectively, and the ground and reference were placed on the earlobes (A1 and A2). This modification was made to provide additional coverage over the frontal cortex. The EOG channels were arranged such that PO9 and PO10 were positioned superior and inferior to the right eye, respectively, while TP9 and TP10 were placed on the left and right temples on the sides of the face, respectively.
In preparation for the experiments, subjects were asked to refrain from using products in their hair that may increase the impedance at the scalp/electrode interface (e.g., conditioner, hair gel, etc.). Prior to donning the cap, the skin on the face around the eyes, the temples, and the earlobes were gently cleaned with alcohol wipes to remove any dirt and skin oils. The cap was aligned on the head such that the FP1 and FP2 were 10% of the distance from the nasion to the inion along the midsaggital plane, and electrode Cz as at the vertex of the head. After donning the cap, a conductive electrolyte gel was applied between the electrode tips and the scalp to reduce the interface impedance. The impedance was maintained below 50 kΩ and in most cases reduced to below 20 kΩ. The channel impedances were recorded prior to the start of the experiment. After completing the gelling process, reference markers were placed on the nasion and left/right pre-auricular points to identify anatomical landmarks. A 3D scanning system (BrainVision Captrak, Brain Products GmbH, Germany) was used to record the 3D electrode positions on the scalp. Finally, the reference markers were removed, and a light surgical mesh was placed over electrodes to fix their location on the scalp and to mitigate the potential influence of motion artifacts during the walking tasks.
Surface EMG signals were recorded using active bipolar electrodes (fixed electrode distance of 20 mm) at 1000 Hz (SX230 sensors and DataLOG MWX8, Biometrics Ltd, Newport, UK). The data was stored on an onboard micro SD card and simultaneously streamed to a PC for visualization and data monitoring using the Biometrics Analysis Software (Biometrics Ltd, Newport, UK). Surface EMG was recorded from six sites bilaterally: tibialis anterior, gastrocnemius, rectus femoris, vastus lateralis, bicep femoris longus, and the semitendinosus. The individual sensor locations were estimated based on limb measurements and anatomical landmarks and validated using recommended clinical tests (i.e., asking subjects to perform a movement and palpating the muscle cite to identify a contraction)40. The recording sites were shaved to remove any hair and subsequently cleaned with an alcohol solution and allowed to dry. The sensors were then fixed to the skin using a double-sided adhesive tape (designed for specific sensor as not to obstruct electrodes). The signals were checked for quality and sensor locations were adjusted as necessary. In cases where the signal was persistently noisy, even after skin preparation, an abrasive skin preparation gel was applied to the cite to remove dead cells and reduce the impedance. The signals were referenced to an electrode located on the skin at the bony prominence created by the styloid process of the ulna. The muscle recording sites were identified published guidelines.
The subjects were instrumented with 17 wireless inertial measurement units (Xsens MVN, Xsens North America Inc., Culver City, CA) for full body motion capture. Each unit was positioned on the body according the manufacturer’s instructions, which included the following locations on the body: (1) head, (2) sternum, (3,4) L/R scapula, (5,6) L/R mid-upper arm, (7,8) L/R mid-forearm, (9,10) L/R hand, (11) L5 spine, (12,13) L/R mid-thigh, (14,15) L/R mid-shank, and (16,17) L/R foot. After donning the 17 IMUs, the system was calibrated while the subject stood in a neutral posture with their hands by their side. The subject was then asked to move about the experimental space, and the accuracy of the movements was validated by visual inspection. Additionally, the magnetic field was continuously monitored to ensure minimal interference and data corruption. The data were recorded at either 30 or 60 Hz (indicated in data) using the MVN Studio software (version 4.4, Xsens North America Inc., Culver City, CA). Information regarding the coordinate systems and algorithms implemented by the software can be found in Roetenberg et al.41
The data were time synchronized using a custom hardware trigger and aligned using MATLAB R2016a (The Mathworks Inc., Natick, MA).
Experimental Protocol
The experiments were conducted on a custom-built gait course (shown in Fig. 1) incorporating five steady locomotion modes: level ground walking (LW), stair descent (SD), stair ascent (SA), ramp descent (RD), and ramp ascent (RA). Prior to beginning the walking trials, the subjects were asked to stand quietly with their eyes open faced away from the gait course (facing a blank wall). One minute of EEG and EMG were recorded to establish a baseline period of brain and muscle activity. Motion capture was not recorded during this period. For the walking trials, the subjects were asked to walk at their preferred speed and to minimize excessive head movements, eye blinks, and jaw movements (e.g., talking, clenching). The subjects began level walking, descended the ramp, gradually turned and transitioned to level ground walking, ascended the staircase, and came to a stop at the end of the stair platform (forward direction). The subject would complete a 180° turn, descend the stairs, transition to level ground, ascend the ramp, and come to rest (reverse direction). Ambulation in both the forward and reverse direction of the course was considered one complete test trial (Fig. 1). The subjects were asked to complete the walking trials in ten blocks of two trials. This was done to allow subjects time to rest between trials if needed and to minimize any potential data loss due to hardware malfunction. The subjects completed an average of twenty trials in approximately ten blocks.
Data Records
All published data are de-identified and subjects gave written informed consent for their data to be openly shared on a credible public data repository. All data files are available from FigShare (Data Citation 1) and have been made available under the terms of Attribution 4.0 International Creative Commons License (http://creativecommons.org/licenses/by/4.0/). The data are archived in a single file set and organized with the following naming convention:
where xx is the subject number (01,02,…,10), yy is the trial number (00, 01, 02,…), abc is the data type (eeg, emg, kin, chanlocs, impedance),.ext is the file format (.mat,.bvct), and the bold text is fixed for all files. It should be noted that the chanlocs, impedance, and emgsensitvity data are recorded once at the beginning of the session, so they will not contain the -T yy entry in the file name. The trials start at 00 indicating the rest, or baseline, trial; all trials thereafter (01, 02, 03,…) are locomotion trials.
-chanlocs.bvct
The -chanlocs.bvct is an XML format file that contains the digitized EEG electrodes locations in 3D space. The data were collected using the BrainVision Captrak software (Brain Products). The file includes the EEG cap size (circumference of the head in cm), the subject’s head shape (round or oval), and the Cartesian/polar coordinates for each electrode (x, y, z, θ, φ, r).
-impedance.mat
The -impedance.mat file contains a structure with two fields (described in Table 1; variable name: impedance).
-eeg.mat
The EEG data are stored in a structure containing two fields (described in Table 2; variable name: eeg). The data stored in eeg.rawdata are raw and unfiltered data without the four EOG channels, which have been moved to eeg.eogdata. In addition to the raw data, the processed data used in ref. 36 have been provided. The data were first high-pass filtered using a fourth-order zero-phase Butterworth filter at 0.01 Hz. Next, channels with a standard deviation greater than 1000 μV or a kurtosis more than five standard deviations from the mean kurtosis of all channels were rejected (using pop_rejchan.m from the EEGLAB toolbox42 for MATLAB). The retained channels were referenced to the common average. Next, we applied artifact subspace reconstruction (ASR)43 on a sliding window of 500 ms to remove corrupted sections with high amplitude bursts (burst criterion=3). Finally, Infomax independent component analysis (ICA) was applied to decompose data into independent sources. Only the ICA sphering and weight matrices are provided in the dataset to reduce size, however, the ICA activations can be easily computed as ICA weights× ICA sphering matrix × processed data. It is important for anyone using these datasets to consider the processing pipeline in detail, as EEG processing can be highly application specific. Otherwise, the raw data should be used to process the data accordingly.
-emg.mat
The EMG data are stored in a structure with two fields (described in Table 3; variable name: emg), each corresponding to a specific leg (emg.left, emg.right).
The channels for both correspond to the following muscles (in the order of the rows from top to bottom): tibialis anterior (TA), medial gastrocnemius (GAST), vastus lateralis (VL), rectus femoris (RF), semitendinosus (SEM), biceps femoris longus (BFL).
-emgsensitivity.mat
The -emgsensitivity.mat file contains a structure with two fields (described in Table 4; variable name: emgsensitivity).
-kin.mat
The -kin.mat file contains a structure (described in Table 5 (available online only); variable name: kin) with the full body motion capture data. The structure contains theee fields, .srate, .setup and .data, with the latter two containing numerous subfields. Table 5 (available online only) breaks down the information contained in each of the fields and subfields.
Technical Validation
Data synchronization
This multi-modal dataset contains recordings from multiple devices/software; thus, external hardware was required for proper synchronization. The hardware consisted of a push-button connected to a 5 V power source. Each system was connected via its respective cable connector. Digital pulses were placed within the data at the beginning and end of each trial and aligned in post processing. The provided data are aligned but in otherwise raw form. This allows future users to process the raw data as desired (e.g., filtering). An example subset of the time-synchronized data is shown in Fig. 2a.
(a) The timeseries EOGH (horizontal) and EOGV (vertical) are computed as bipolar signals for the horizontal and vertical EOG channels, respectively. (b) Impedance values (kΩ) of the 60-channel EEG for each of the ten subjects just prior to beginning the experiment. (c) The total α power (dB) in all the EEG channels (60 electrodes) during each of the conditions (standing, LW, SA, from left to right) is shown above the timeseries plot. Gradual increases in α band desynchronization can be observed between as the subject transitions from standing, to level ground walking, to stair walking, respectively.
EEG, Captrak, and Impedance data
The EEG setup was rigorously prepared to minimize potential artifacts and improve the signal-to-noise ratio (complete details of this are included in the methods). The impedance for each electrode was reduced to below 50 kΩ prior to beginning the experiment (shown in Fig. 2b).
Event related desynchronization (reduction in power or ERD) in the α and β band during movement onset has been widely documented in the literature44,45. This characteristic behavior is observed in our data. As shown in Fig. 2, there is a clear reduction in α power when the subject is walking on level ground when compared to standing. Furthermore, there is an additional reduction in α power during stair ascent when compared to level ground walking. The α power (dB) scalp maps shown in Fig. 2c were computed using the following steps: (1) band pass filtered between 0.3–50 Hz using a fourth order Butterworth filter; (2) referenced to the common average (using reref.m in MATLAB); (3) the power spectral density estimate was computed for each channel using the Thomson multitaper method (pmtm.m in MATLAB; 3.5 discrete prolate spheroidal sequences); and the average log power in the α band (8–12 Hz) for each channel was projected onto the scalp using the topoplot.m function in the EEGLAB toolbox for MATLAB42.
EMG data
The quality of the lower-limb EMG signals was assessed prior to the beginning of the experiment. The gains were adjusted so that none of the channels were subject to digital clipping. As shown in Fig. 2a, the muscle activations can be clearly seen during the walking conditions (LW and SA). Furthermore, the activations of antagonistic muscles (e.g., TA/GAST and VL/BFL) are approximately 180° out of phase as expected during locomotor tasks.
Kinematic data
The motion capture system was carefully setup according to the manufacturer’s instructions. After donning the 17 IMUs, the system was calibrated while the subject stood in a neutral posture with their hands by their side. The subject was then asked to move about the experimental space, and the accuracy of the movements was validated by visual inspection. Finally, the magnetic field was continuously monitored to ensure minimal interference and data corruption. Figure 3 shows the hip, knee, and ankle joint angles for all ten subjects during locomotion on level ground (left column) and stairs (right column).
Additional information
How to cite this article: Brantley, J. A. et al. Full body mobile brain-body imaging data during unconstrained locomotion on stairs, ramps, and level ground. Sci. Data 5:180133 doi: 10.1038/sdata.2018.133 (2018).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
References
Rossignol, S., Dubuc, R. & Gossard, J. P. Dynamic sensorimotor interactions in locomotion. Physiol. Rev 86, 89–154 (2006).
Cevallos, C. et al. Oscillations in the human brain during walking execution, imagination and observation. Neuropsychologia 79, 223–232, 10.1016/j.neuropsychologia.2015.06.039 (2015).
Cheron, G. et al. From Spinal Central Pattern Generators to Cortical Network: Integrated BCI for Walking Rehabilitation. Neural Plast., 375148, 10.1155/2012/375148 (2012).
Hamacher, D., Herold, F., Wiegel, P., Hamacher, D. & Schega, L. Brain activity during walking: A systematic review. Neurosci. Biobehav. Rev. 57, 310–327, 10.1016/j.neubiorev.2015.08.002 (2015).
Beisteiner, R., Hollinger, P., Lindinger, G., Lang, W. & Berthoz, A. Mental representations of movements. Brain potentials associated with imagination of hand movements. Electroencephalogr. Clin. Neurophysiol. 96, 183–193 (1995).
Dechent, P., Merboldt, K. D. & Frahm, J. Is the human primary motor cortex involved in motor imagery? Brain Res. Cogn. Brain Res 19, 138–144 (2004).
Jackson, P. L., Lafleur, M. F., Malouin, F., Richards, C. L. & Doyon, J. Functional cerebral reorganization following motor sequence learning through mental practice with motor imagery. Neuroimage 20, 1171–1180 (2003).
Jahn, K. et al. Imaging human supraspinal locomotor centers in brainstem and cerebellum. Neuroimage 39, 786–792 (2008).
Jahn, K. et al. Brain activation patterns during imagined stance and locomotion in functional magnetic resonance imaging. Neuroimage 22, 1722–1731 (2004).
Miyai, I. et al. Cortical mapping of gait in humans: a near-infrared spectroscopic topography study. Neuroimage 14, 1186–1192 (2001).
Suzuki, M., Miyai, I., Ono, T. & Kubota, K. Activities in the frontal cortex and gait performance are modulated by preparation. An fNIRS study. Neuroimage 39, 600–607 (2008).
Suzuki, M. et al. Prefrontal and premotor cortices are involved in adapting walking and running speed on the treadmill: an optical imaging study. Neuroimage 23, 1020–1026 (2004).
Rutkove, S. in The Clinical Neurophysiology Primer (eds Blum Andrew, S. & Seward, B. Rutkove ) Ch. 4, 43–53 (Humana Press, 2007).
van den Broek, S. P., Reinders, F., Donderwinkel, M. & Peters, M. J. Volume conduction effects in EEG and MEG. Electroencephalogr. Clin. Neurophysiol 106, 522–534 (1998).
Bradford, J. C., Lukos, J. R. & Ferris, D. P. Electrocortical activity distinguishes between uphill and level walking in humans. J. Neurophysiol 115, 958–966 (2016).
Bulea, T. C., Kim, J., Damiano, D. L., Stanley, C. J. & Park, H.-S. Prefrontal, posterior parietal and sensorimotor network activity underlying speed control during walking. Front. Hum. Neurosci. 9, 247 (2015).
Castermans, T., Duvinage, M., Cheron, G. & Dutoit, T. About the cortical origin of the low-delta and high-gamma rhythms observed in EEG signals during treadmill walking. Neurosci. Lett. 561, 166–170 (2014).
Gramann, K., Gwin, J. T., Bigdely-Shamlo, N., Ferris, D. P. & Makeig, S. Visual Evoked Responses During Standing and Walking. Front. Hum. Neurosci. 4, 202 (2010).
Gwin, J. T., Gramann, K., Makeig, S. & Ferris, D. P. Electrocortical activity is coupled to gait cycle phase during treadmill walking. Neuroimage 54, 1289–1296 (2011).
Seeber, M., Scherer, R., Wagner, J., Solis-Escalante, T. & Muller-Putz, G. R. EEG beta suppression and low gamma modulation are different elements of human upright walking. Front. Hum. Neurosci. 8, 485 (2014).
Seeber, M., Scherer, R., Wagner, J., Solis-Escalante, T. & Müller-Putz, G. R. High and low gamma EEG oscillations in central sensorimotor areas are conversely modulated during the human gait cycle. Neuroimage 112, 318–326 (2015).
Severens, M., Nienhuis, B., Desain, P. & Duysens, J. in Conf. Proc. IEEE Eng. Med. Biol. Soc. 2764–2767, doi:10.1109/EMBC.2012.6346537 (2012).
Wagner, J. et al. Level of participation in robotic-assisted treadmill walking modulates midline sensorimotor EEG rhythms in able-bodied subjects. Neuroimage 63, 1203–1211 (2012).
Wagner, J., Solis-Escalante, T., Scherer, R., Neuper, C. & Müller-Putz, G. It's how you get there: walking down a virtual alley activates premotor and parietal areas. Front. Hum. Neurosci 8, 93 (2014).
Gwin, J. T., Gramann, K., Makeig, S. & Ferris, D. P. Removal of movement artifact from high-density EEG recorded during walking and running. J. Neurophysiol 103, 3526–3534 (2010).
Petersen, T. H., Willerslev-Olsen, M., Conway, B. A. & Nielsen, J. B. The motor cortex drives the muscles during walking in human subjects. J. Physiol 590, 2443–2452 (2012).
Lau, T. M., Gwin, J. T. & Ferris, D. P. Walking reduces sensorimotor network connectivity compared to standing. J. Neuroeng. Rehabil 11, 14 (2014).
Nathan, K. & Contreras-Vidal, J. L. Negligible Motion Artifacts in Scalp Electroencephalography (EEG) During Treadmill Walking. Front. Hum. Neurosci 9, 708 (2015).
Langer, N. et al. A resource for assessing information processing in the developing brain using EEG and eye tracking. Sci. Data 4, 170040 (2017).
Mouček, R., Vařeka, L., Prokop, T., Štěbeták, J. & Brůha, P. Event-related potential data from a guess the number brain-computer interface experiment on school children. Sci. Data 4, 160121 (2017).
Schauer, G. et al. Fractionation of parietal function in bistable perception probed with concurrent TMS-EEG. Sci. Data 3, 160065 (2016).
Wakeman, D. G. & Henson, R. N. A multi-subject, multi-modal human neuroimaging dataset. Sci. Data 2, 150001 (2015).
Luciw, M. D., Jarocka, E. & Edin, B. B. Multi-channel EEG recordings during 3,936 grasp and lift trials with varying weight and friction. Sci. Data 1, 140047 (2014).
Brantley, J. A. et al. in Conf. Proc. IEEE Eng. Med. Biol. Soc. 5729-5732, doi:10.1109/EMBC.2016.7592028 (2016).
Luu, T. P., Brantley, J. A., Zhu, F. & Contreras-Vidal, J. L. in Conf. Proc. IEEE Eng. Med. Biol. Soc. 1913-1916, doi:10.1109/embc.2017.8037222 (2017).
Luu, T. P., Brantley, J. A., Nakagome, S., Zhu, F. & Contreras-Vidal, J. L. Electrocortical correlates of human level-ground, slope, and stair walking. PLoS ONE 12, e0188500 (2017).
Luu, T. P., Brantley, J. A., Zhu, F. & Contreras-Vidal, J. L in 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC). 2437-2441, doi:10.1109/SMC.2017.8122988 (2017).
Nakagome, S., Luu, T. P., Brantley, J. A. & Contreras-Vidal, J. L in 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC). 3175-3178, doi:10.1109/SMC.2017.8123116 (2017).
Brantley, J. A., Luu, T. P., Nakagome, S. & Contreras-Vidal, J. L in 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC). 1705-1709, doi:10.1109/SMC.2017.8122861 (2017).
Cram, J. R. Cram's introduction to surface electromyography. 2nd edn (Jones & Bartlett Learning, 2011).
Roetenberg, D., Luinge, H. & Slycke, P. Xsens MVN: full 6DOF human motion tracking using miniature inertial sensors (Xsens Motion Technologies BV, Tech. Rep, 2009).
Delorme, A. & Makeig, S. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 134, 9–21 (2004).
Mullen, T. et al. in Conf. Proc. IEEE Eng. Med. Biol. Soc. 2184-2187, doi:10.1109/embc.2013.6609968 (2013).
Neuper, C., Wörtz, M., Pfurtscheller, G. in Prog. Brain Res. Vol. Volume 159 (eds Neuper Christa & Klimesch Wolfgang ) 211–222 (Elsevier, 2006).
Pfurtscheller, G. & Lopes da Silva, F. H. Event-related EEG/MEG synchronization and desynchronization: basic principles. Clin. Neurophysiol. 110, 1842–1857 (1999).
Data Citations
Brantley, J., Luu, T. P., Zhu, F., Nakagome, S., & Contreras-Vidal, J. L. Figshare https://doi.org/10.6084/m9.figshare.5616109.v5 (2018)
Acknowledgements
This research is partly supported by NSF award IIS-1302339 and an NIH F99 Predoctoral Fellowship (1F99NS105210-01) to JAB. The authors would like to acknowledge Dr. Beom-Chan Lee for generously sharing the Xsens MVN system along with the Center for Neuromotor and Biomechanics Research (CNBR) for permitting use of other equipment and space. We also would like to thank Dr. Recep A Ozdemir for his assistance in data collection.
Author information
Authors and Affiliations
Contributions
J.B. wrote the manuscript and led the data collection. T.P.L. and S.N. contributed to the manuscript and data collection. F.Z. contributed to the data collection. J.L.C.V. designed the experiment, supervised the project, and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
ISA-Tab metadata
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files made available in this article.
About this article
Cite this article
Brantley, J., Luu, T., Nakagome, S. et al. Full body mobile brain-body imaging data during unconstrained locomotion on stairs, ramps, and level ground. Sci Data 5, 180133 (2018). https://doi.org/10.1038/sdata.2018.133
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/sdata.2018.133
This article is cited by
-
A Non-Laboratory Gait Dataset of Full Body Kinematics and Egocentric Vision
Scientific Data (2023)
-
Development and Uncertainty Assessment of Low-Cost Portable EMG Acquisition Module
MAPAN (2023)
-
Lower-limb kinematics and kinetics during continuously varying human locomotion
Scientific Data (2021)
-
Mobile BCI dataset of scalp- and ear-EEGs with ERP and SSVEP paradigms while standing, walking, and running
Scientific Data (2021)
-
GaitRec, a large-scale ground reaction force dataset of healthy and impaired gait
Scientific Data (2020)