Abstract
Structural brain data is key for the understanding of brain function and networks, i.e., connectomics. Here we present data sets available from the ‘atlasing of the basal ganglia (ATAG)’ project, which provides ultra-high resolution 7 Tesla (T) magnetic resonance imaging (MRI) scans from young, middle-aged, and elderly participants. The ATAG data set includes whole-brain and reduced field-of-view MP2RAGE and T2*-weighted scans of the subcortex and brainstem with ultra-high resolution at a sub-millimeter scale. The data can be used to develop new algorithms that help building high-resolution atlases both relevant for the basic and clinical neurosciences. Importantly, the present data repository may also be used to inform the exact positioning of electrodes used for deep-brain-stimulation in patients with Parkinson’s disease and neuropsychiatric diseases.
Design Type(s) | parallel group design • observation design |
Measurement Type(s) | nuclear magnetic resonance assay |
Technology Type(s) | MRI Scanner |
Factor Type(s) | age |
Sample Characteristic(s) | Homo sapiens • brain |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
Large collaborative projects between scientific groups spread around the world are aimed to increase our understanding of the human brain. Large human connectome studies1–3 are in place working to clarify the connectivity within the human brain using a multi-modal approach ranging from structural brain imaging to genetics (http://www.humanconnectomeproject.org). However, to fully understand the connectivity of the brain, we need a higher level of anatomical detail than currently available. The lack of knowledge about small brain structures, especially subcortical structures, is reflected by their absence from brain atlases currently available for MRI research4,5. A comparison of subcortical grey matter structures depicted in standard MRI-atlases with the structures defined in the Federative Community on Anatomical Terminology6 yielded an overlap of only seven percent. One important explanation for this discrepancy is the absence of ultra-high resolution MRI data allowing the direct visualization of small nuclei in the subcortex. A second important reason is the lack of automated analytical protocols available for MRI-data segmentation, with the resulting necessity of laborious studies performed by trained anatomists for the identification of subcortical brain areas. Thirdly, besides the lack of anatomical knowledge, there is no information about age-related changes in, e.g., volume or location of subcortical structures.
Recent exciting advancements in the field of ultra-high resolution magnetic resonance imaging at 7 Tesla (or higher) allow in vivo neuroimaging of the human brain with unprecedented anatomical detail7–11. Here we share information of a multi-modal data set of three different groups of young, middle-aged, and elderly participants who were scanned with a 7 T MRI scanner. The data sets contain three different age groups and can be used to investigate anatomical changes due to healthy aging. The data sets have already been used to create probabilistic atlas maps including the striatum, globus pallidus interna and externa, the substantia nigra, the subthalamic, and the red nucleus. All probabilistic atlas maps are available online (https://www.nitrc.org/projects/atag/ and http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/Atlases). In addition to the manual segmentations, the data can be used to develop new algorithms that help building high-resolution subcortical brain atlases that can be directly applied in both the basic and clinical neurosciences. Finally, the data can be used to guide the exact positioning of electrodes relevant for deep-brain-stimulation often used in patients with Parkinson’s disease and neuropsychiatric diseases12–14.
Methods
Participants
For the acquisition of the structural brain scans, 30 young participants (14 females) with mean age 23.8 (s.d. 2.3), 14 middle-aged (7 females) with mean age 52.5 (s.d. 6.6), and 10 elderly (3 females) with mean age 69.6 (s.d. 4.6) were included (Table 1). All participants had normal or corrected-to-normal vision, and none of them suffered from neurological, psychiatric, or somatic diseases. All subjects were right-handed, as confirmed by the Edinburgh Inventory15. The study was approved by the local ethics committee at the University of Leipzig, Germany. All participants gave their written informed consent prior to scanning and received a monetary compensation.
Scan parameters
The structural data were acquired using a 7 T Siemens Magnetom MRI scanner using a 24-channel head array Nova coil (NOVA Medical Inc., Wilmington MA) and consisted of three sequences: a whole-brain MP2RAGE, a MP2RAGE covering a smaller slab16,17, and a multi-echo 3D FLASH18. The whole-brain MP2RAGE had 240 sagittal slices with an acquisition time of 10:57 min (repetition time (TR)=5,000 ms; echo time (TE)=2.45 ms; inversion times TI1/TI2=900/2,750 ms; flip angle=5°/3°; bandwidth=250 Hz/Px; voxel size=(0.7 mm)3; Table 2 (available online only)). The MP2RAGE slab consisted of 128 slices with an acquisition time of 9:07 min (TR=5,000 ms; TE=3.71 ms; TI1/TI2=900/2,750 ms; flip angle=5°/3°; bandwidth=240 Hz/Px; voxel size=(0.6 mm)3; Table 3 (available online only)). The FLASH slab consisted of 128 slices with an acquisition time of 17:18 min (TR=41 ms and three different echo times (TE): 11.22/20.39/29.57 ms; flip angle=14°; bandwidth=160 Hz/Px; voxel size=(0.5 mm)3; Table 4 (available online only)). Both slab sequences consisted of axial slices tilted −23 degrees to the true axial plane in scanner coordinates. This angle in combination with the used field of view ensured that the entire Basal Ganglia were scanned. To get a better inversion of the magnetization in the lower parts of the brain (e.g., the Cerebellum), a TR-FOCI inversion pulse was implemented in the MP2RAGE sequence16.
Unless indicated otherwise, all MRI data files were converted from DICOM to NIfTI format using an in-house dicom-to-nifti converter. This linux compatible converter is available via https://github.com/isis-group/isis.
Scan volumes
The MP2RAGE sequence results in four different volumes for each subject: INV1, INV2, UNI and T1. The INV1 volume reflects the gradient echo sequence with an inversion time of 900 ms. The INV2 volume reflects the gradient echo sequence with an inversion time of 2,750 ms. The UNI volume is the combined volume of the two inversion times. Finally, the T1 volume is a T1 estimation map derived from the two inversion times (Marques et al.17). The FLASH sequence results in two different volumes per echo time per subject resulting in nine different volumes in total. Besides the standard T2* weighted magnitude image, the phase images are also provided and can be used to calculate susceptibility weighted images as well as quantitative susceptibility maps (e.g., Deistung et al.19).
Data processing
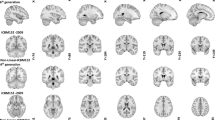
All structural scans were anonymized by zeroing out the voxels in the vicinity of the facial surface, teeth, and auricles following a similar procedure as described by Hanke et al.20 All data were reoriented to the standard MNI space using the fslreorient2std tool as implemented in fslutils 5.0.2 (Figure 1).
Three different age groups were structurally scanned using a 7 T MRI scanner. Data acquisition was done in a single imaging session that lasted for approximately 37 min. This resulted in three different datasets: a whole brain T1-weighted MP2RAGE volume; a slab T1-weighted MP2RAGE volume, and a T2*-weighted flash volume. All structural data was anonymized and reoriented to standard MNI orientation (7 T MRI photo courtesy of Andreas Döring).
Data Records
All data records listed in this section are available from NITRIC (Data Citation 1) or Dryad (Data Citation 2). A README file with a detailed description of the content of all downloads is available in Dryad. Additional material and information are also provided in Data Citation 1 and Data Citation 2.
Unless noted otherwise, all MRI data files were converted from DICOM to NIfTI format using an in-house dicom-to-nifti converter. In order to de-identify data, information on centre-specific study and participant codes have been removed using an automated procedure. All human participants were given sequential integer IDs.
Technical Validation
Motion artifacts
In line with Gedamu et al.21, motion artifacts in the structural volumes were estimated by calculating the noise ratio between the phase encoding direction and read direction outside of the brain. Two ROIs of +/−1,225 mm2 was drawn in the sagittal plane; 5 mm lateral of the skull, and in the coronal plane; 5 mm anterior of the skull, in the magnitude image of the second inversion time of the MP2RAGE sequence and FLASH sequences. The sagittal ROI corresponds to the read direction for the MP2RAGE whole brain and phase encoding direction for the MP2RAGE and FLASH slab, whereas the coronal ROI corresponds to the phase encoding direction for the MP2RAGE whole brain and read direction for the MP2RAGE and FLASH slab. The mean signal was extracted from both ROI’s and the mean phase encoding direction signal was divided by the mean read direction signal. The closer this ratio is to 1, the less motion artifacts are present. Following Gedamu et al.21, we estimated that any ratio below 2 reflects little to no motion artifacts (see Figure 2 for an example of the data quality).
Two axial images of the three acquired datasets are displayed for a representative young subject. Only a few of the easily identifiable structures have been labeled. Note that not all structures are equally well visibly in the T1-weighted volumes compared to the T2*-weighted volume and argue for the need of multi sequence acquisition when interested in subcortical structures.
One sided t-tests were conducted to test whether any of the groups showed significant motion artifacts in any of the sequences. All ratios per sequence and age group were significantly lower than 2 (MP2RAGE whole-brain: young (t(29)=−17.93, P<0.001); middle-aged (t(13)=−5.44, P<0.001); elderly (t(8)=−7.19, P<0.001), MP2RAGE slab: young (t(29)=−35.06, P<0.001); middle-aged (t(13)=−23.43, P<0.001); elderly (t(8)=−13.33, P<0.001), FLASH echo 1: young (t(29)=−3.74, P<0.001); middle-aged (t(13)=−17.68, P<0.001); elderly (t(8)=−16.97, P<0.001), FLASH echo 2: young (t(29)=−6.88, P<0.001); middle-aged (t(13)=−14.88, P<0.001); elderly (t(8)=−6.31, P<0.001), FLASH echo 3: young (t(29)=−10.36, P<0.001); middle-aged (t(13)=−6.23, P<0.001); elderly (t(8)=−19.53, P<0.001); Table 5 (available online only)).
There was no main effect of age on motion for the MP2RAGE whole brain (F(2,50)=1.29, P=0.29) or MP2RAGE slab (F(2,50)=0.8, P=0.44). There was a main effect of age and echo time on motion for the FLASH sequence (age: F(2,147)=4.97, P=0.008, echo time: F(2,147)=10.45, P<0.001). Post-hoc testing showed that the young had significantly more motion artifacts than both the middle-aged and elderly (young versus middle-aged: t(103.18)=5.61, P<0.001, young versus elderly: t(79.03)=5.25, P<0.001) whereas the middle-aged and elderly did not differ significantly (t(65.73)=−0.59, P=1.0). Post-hoc testing showed that the first echo time had significantly less motion artifacts than both the second and third echo time (first echo versus second echo: t(90)=−3.29, P=0.003, first echo versus third echo: t(82.17)=−3.77, P=0.001) whereas the second and third echo time did not differ significantly (t(101.66)=−0.72, P=0.92). All post-hoc testing was Bonferroni corrected at an alpha of 0.05.
Signal to noise ratio
To estimate the Signal to Noise Ratio (SNR), the mean signal from an axial slice just above the corpus callosum was divided by the standard deviation of the signal in the read direction ROI both in the magnitude image of the second inversion time of the MP2RAGE sequence and FLASH sequences. To improve the estimation of noise a Rician correction was applied22. As this is still an approximation of the true SNR, the term SNRapprox. is used. For the three different sequences there was a main effect of age on SNRapprox. (MP2RAGE whole brain: F(2,50)=48.3, P<0.001; MP2RAGE slab: F(2,50)=5.94, P=0.005; FLASH: F(2,147)=6.90, P=0.001). Additionally there was a main effect of echo time on SNRapprox. (F(2,147)=11.75, P<0.001).
Post-hoc testing showed that for the MP2RAGE whole brain, the young had a significantly higher SNRapprox than both the middle-aged and elderly (young versus middle-aged: t(33.72)=8.87, P<0.001; young versus elderly: t(18.37)=8.41, P<0.001) whereas the middle-aged and elderly did not differ significantly (t(17.61)=0.46, P=1.0). A similar pattern was found for the MP2RAGE slab. The young had a significantly higher SNRapprox than both the middle-aged and elderly (young versus middle-aged: t(24.8)=2.86, P=0.017; young versus elderly: t(17.46)=2.92, P=0.019) whereas the middle-aged and elderly did not differ significantly (t(20.56)=−0.10, P=1.0). The young had a significantly higher SNRapprox in the FLASH sequence than the middle-aged (t(70.80)=3.35, P=0.003) but did not differ from the elderly (t(36.31)=0.16, P=1.0). The middle-aged and elderly did not differ in SNRapprox for the FLASH sequence (t(51.87)=−2.16, P=0.071). Post-hoc testing showed that the first echo time had significantly more SNRapprox than both the second and third echo time (first echo versus second echo: t(97.2)=4.89, P<0.001, first echo versus third echo: t(88.4)=8.05, P<0.001). The second echo time had significantly higher SNRapprox than the third echo time (t(100.91)=3.42, P=0.002). All post-hoc testing was Bonferroni corrected at an alpha of 0.05 (Table 6 (available online only)).
In addition to the SNRapprox. calculation and the noise ratio between the phase encoding direction and read direction, the scans were visually inspected by two independent researchers. The FLASH magnitude scans were checked for ghosting, wrapping, or shading artifacts. The MP2RAGE UNI scans were checked for ghosting, wrapping, shading, and the presence of ‘zebra stripe’ artifacts. Finally the MP2RAGE T1 scans were checked for ghosting, wrapping, shading, the presence of ‘zebra stripes’, and CSF clipping artifacts where ‘1’ corresponds to not present at all and ‘5’ corresponds to severely present.
Ghosting artifacts are generally caused by motion and appear as a ‘ghost’ image of the brain in phase encoding direction. Wrapping artifacts are usually caused by anatomical features protruding outside of the imaged field of view but still within the sensitive volume of the RF coil. Shading artifacts were defined as a non-homogenous intensity throughout the entire brain. Zebra stripes were defined as well defined alternating black and white stripes present in the brain. Finally, CSF clipping artifacts were defined as the voxels in the CSF that have a signal dropout and appear black (McRobbie et al., 2006).
The mean rating for each scale for each checked volume is given in Table 7 (available online only). Volumes that had a higher rating on that quality check than the rest of the age group based on the +/−1.5* interquartile range are highlighted with an asterisk.
As a result of the scan parameters of the MP2RAGE sequence, a number of participants show T1 clipping artifacts in the T1 map located in the CSF. This is indicated in Table 7 (available online only). Note that these clipping artifacts do not affect the T1 values reported in the grey and white matter tissue.
Usage Notes
The procedures we employed in this study resulted in a dataset that is highly suitable for automated processing. Data are shared in documented standard formats, such as NIfTI or plain text files, to enable further processing in arbitrary analysis environments with no imposed dependencies on proprietary tools. All processing performed on the released data article were produced by open-source software on standard computer workstations.
References
References
Biswal, B. B. et al. Toward discovery science of human brain function. Proc. Natl Acad. Sci. 107, 4734–4739 (2010).
Sporns, O., Tononi, G. & Kötter, R. The Human Connectome: A structural description of the human brain. PLoS Comp. Biol. 1, e42 (2005).
Van Essen, D. C. et al. The Human Connectome Project: A data acquisition perspective. NeuroImage 62, 2222–2231 (2012).
Evans, A. C., Janke, A. L., Collins, D. L. & Baillet, S. Brain templates and atlases. NeuroImage 62, 911–922 (2012).
Alkemade, A., Keuken, M. C. & Forstmann, B. U. A perspective on terra incognita: uncovering the neuroanatomy of the human subcortex. Frontiers in Neuroanatomy 7, 40 (2013).
Federative Committee on Anatomical Terminology. Terminologia Anatomica 1–292 (Thieme Stuttgart, 1998).
Lenglet, C. et al. Comprehensive in vivo mapping of the human basal ganglia and thalamic connectome in individuals using 7T MRI. PloS one 7, e29153 (2012).
Keuken, M. C. et al. Quantifying inter-individual anatomical variability in the subcortex using 7T structural MRI. NeuroImage 94, 1–7 (2014).
Cho, Z. H. et al. New brain atlas—Mapping the human brain in vivo with 7.0 T MRI and comparison with postmortem histology: Will these images change modern medicine? Int. J. Imag. Syst. Tech. 18, 2–8 (2008).
Turner, R. in High-Field MR Imaging (Springer, 2011).
Bazin, P.-L. et al. A computational framework for ultra-high resolution cortical segmentation at 7Tesla. NeuroImage 1–9 (2013).
Beisteiner, R. et al. Clinical fMRI: Evidence for a 7T benefit over 3T. NeuroImage 57, 1015–1021 (2011).
Cho, Z. H. et al. Direct visualization of deep brain stimulation targets in Parkinson disease with the use of 7-tesla magnetic resonance imaging. J. Neurosurg. 113, 1–9 (2010).
Abosch, A., Yacoub, E., Ugurbil, K. & Harel, N. An assessment of current brain targets for deep brain stimulation surgery with susceptibility-weighted imaging at 7-tesla. Neurosurgery 67, 1745–1756 (2010).
Oldfield, R. C. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia 9, 97–113 (1971).
Hurley, A. C. et al. Tailored RF pulse for magnetization inversion at ultrahigh field. Magn. Reson. Med. 63, 51–58 (2009).
Marques, J. P. et al. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage 49, 1271–1281 (2010).
Haase, A., Frahm, J., Matthaei, D., Hanicke, W. & Merboldt, K. D. FLASH imaging. Rapid NMR imaging using low flip-angle pulses. J. Magn. Reson. 67, 258–266 (1986).
Deistung, A. et al. Toward in vivo histology: A comparison of quantitative susceptibility mapping (QSM) with magnitude-, phase-, and R2*-imaging at ultra-high magnetic field strength. NeuroImage 65, 299–314 (2013).
Hanke, M. et al. A high-resolution 7-Tesla fMRI dataset from complex natural stimulation with an audio movie. Sci. Data 1 140003 (2014).
Gedamu, E. L., Collins, D. L. & Arnold, D. L. Automated quality control of brain MR images. J. Magn. Reson. Imaging 28, 308–319 (2008).
Gudbjartsson, H. & Patz, S. The Rician distribution of noisy MRI data. Magn. Reson. Med. 34, 910–914 (1995).
Data Citations
Forstmann, B. U. NITRC (2014) http://www.nitrc.org/projects/atag_mri_scans/
Forstmann, B. U. Dryad (2014) http://doi.org/10.5061/dryad.fb41s
Acknowledgements
We thank Domenica Wilfling and Elisabeth Wladimirov for taking such good care of all our participants. This research line is financially supported by the European Research Council (BUF).
Author information
Authors and Affiliations
Contributions
B.U.F. conceived the study and wrote the manuscript. M.C.K. contributed to the manuscript, performed the technical validation, and visually checked the data. A.S. contributed to the manuscript and performed the technical validation. P.-L.B. contributed to the manuscript and provided conceptual discussion. A.A. contributed to the manuscript and visually checked the data. R.T. provided conceptual discussion and contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interest.
ISA-Tab metadata
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0 Metadata associated with this Data Descriptor is available at http://www.nature.com/sdata/ and is released under the CC0 waiver to maximize reuse.
About this article
Cite this article
Forstmann, B., Keuken, M., Schafer, A. et al. Multi-modal ultra-high resolution structural 7-Tesla MRI data repository. Sci Data 1, 140050 (2014). https://doi.org/10.1038/sdata.2014.50
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/sdata.2014.50
This article is cited by
-
Activation of cerebellum and basal ganglia during the observation and execution of manipulative actions
Scientific Reports (2020)
-
Towards a mechanistic understanding of the human subcortex
Nature Reviews Neuroscience (2017)
-
Effects of aging on \(T_{1}\) T 1 , \(T_{2}^{*}\) T 2 ∗ , and QSM MRI values in the subcortex
Brain Structure and Function (2017)