Abstract
Study design:
Retrospective observational study of bacterial susceptibilities in Veterans with SCI/D as compared to a general patient population.
Objectives:
The purpose of this project was to evaluate the prevalence and susceptibility of bacteria isolated from spinal cord injury and disorder (SCI/D) patients as compared with a general patient population and determine whether a SCI/D-specific antibiogram, a report of bacterial susceptibilities used to guide empiric antibiotic selection, would be a useful stewardship tool.
Setting:
Veterans Affairs Medical Center located in Cook county, IL, USA.
Methods:
Microbiology reports from 1 October 2012 to 30 September 2013 were compiled into a SCI/D-specific antibiogram and compared to a non-SCI/D antibiogram.
Results:
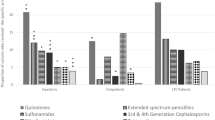
Persons with positive cultures and SCI/D were younger and had a higher Charlson Index as compared to non-SCI/D patients (P<0.0001 for both). Five thousand one hundred and thirty-one unique isolate cultures were evaluated (SCI/D=23.0%). Frequencies of pathogens isolated in SCI/D and non-SCI/D differed. Methicillin-resistant Staphylococcus aureus occurred more frequently in SCI/D (27.8% vs 55.4%; P<0.0001). Gram-negatives had generally lower susceptibilities in SCI/D and a higher frequency of organisms producing extended-spectrum Beta-lactamases (17.6% vs 5.0%; P<0.0001), carbapenem-resistant Enterobacteriaceae (2.4% vs 0.5%; P<0.0001), carbapenem resistance (7.6% vs 2.4%; P<0.0001) and isolates resistant to ⩾3 antibiotic classes (60.7% vs 28.0%; P=0.0001).
Conclusion:
Different pathogens with poorer susceptibilities are isolated in SCI/D. Thus an SCI/D-specific antibiogram reflective of resistance patterns in these patients may increase the appropriateness of empiric antibiotic selection. The frequency of multi-drug resistant organisms in cultures obtained from patients with SCI/D is worrisome.
Similar content being viewed by others
Introduction
Antibiotic resistance increases the likelihood of initial inadequate therapy, which corresponds to an increased mortality, length of stay and health-care costs.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 Antibiograms are facility-specific aggregated reports of bacterial susceptibilities and are recommended as a component of antimicrobial stewardship programs.15 Clinically, the antibiogram is used as a tool to aid providers in making evidence-based decisions when choosing the most appropriate agent for initial empiric antibiotic treatment because they are reflective of the susceptibility of organisms isolated at that facility.16 Thus antibiograms specific to certain patient care areas may allow for identification of resistance issues specific to that population and allow for focused antimicrobial stewardship and infection-control efforts.15, 17, 18 However, less than half of academic medical centers have unit-specific antibiograms.19 As inappropriate initial empiric antibiotic treatment is associated with increased morbidity and mortality, the lack of unit-specific antibiograms may negatively affect clinical outcomes because clinicians have less information available to guide treatment selection.15 Further, previous studies comparing unit-specific antibiograms to hospital-wide antibiograms found significant differences in the susceptibility rates across different units,20, 21 suggesting that use of unit-specific or population-stratified antibiograms might be appropriate.22
Individuals with spinal cord injury or disorder (SCI/D) are at high risk for infections and antimicrobial resistance compared to the general patient population secondary to factors, such as frequent hospitalization, previous antibiotic use, frequent and chronic use of invasive devices and development of pressure ulcers.23, 24 However, data on bacterial resistance in the SCI/D population is scare and use of antibiograms in hospital units caring for these patients is limited.25 The lack of SCI-specific antimicrobial susceptibility data poses a significant limitation to the optimal management of infectious diseases in SCI/D patients, because resistance patterns differ based on patient history, care area and past exposure to health-care services and antimicrobials.26, 27 This leads to potential significant bias in using hospital antibiograms to guide treatment decisions, particularly for complicated patients with SCI/D. Thus the purpose of this project was to evaluate the prevalence and resistance patterns of bacteria isolated from patients with SCI/D as compared to a general patient population and determine whether an SCI/D-specific antibiogram would be valuable for facilities treating these patients.
Materials and methods
Study design, setting and population
A retrospective cross-sectional analysis of microbiology laboratory reports from 1 October 2012 to 30 September 2013 for all cultures (inpatient and outpatient) obtained at a 471-bed Midwestern VA facility were evaluated. The SCI/D cohort at the study facility was identified using an ongoing registry of Veterans with SCI/D maintained by the VA Allocation Resource Center (ARC), which is used to allocate resources to VA medical centers.28 The SCI/D group was compared with the remaining non-SCI/D patients with cultures during the study period. Patient location (inpatient vs outpatient) was defined as the patient location when the culture was taken. This study was reviewed and approved by the local institutional review board.
Microbiology data and definitions
Microbiology data were obtained from the VA’s Corporate Data Warehouse, which includes data on medical encounters occurring in the VA health-care system. Microbiology records were queried to extract all cultures for aerobic and anaerobic bacteria, regardless of the source (for example, blood, urine, sputum). Only the first isolate cultured from a single patient was included unless the susceptibility pattern changed. Consistent with current recommendations for comparing susceptibility results, only those bacteria isolated in ⩾30 non-surveillance cultures were included in this evaluation.29 Extended spectrum beta-lactamase (ESBL) producing Enterobacteriaceae and Klebsiella-producing carbapenemases (KPC) were conducted using broth microdilution and the modified Hodge test, respectively, using standardized Clinical and Laboratory Standards Institute methodology.30
Multi-drug resistant Gram-negatives were defined as isolates with intermediate or resistant susceptibilities to ⩾3 of the following classes/agents: aminoglycosides, ampicillin, aztreonam, antipseudomonal penicillins in combination with a Beta-lactamase inhibitor, carbapenems, cephalosporins, cephamycins, chloramphenicol, colistin, fluoroquinolones, fosfomycin, penicillins in combination with a Beta-lactamase inhibitor, sulfamethoxazole/trimethoprim, tetracyclines, and tigecycline.30 Carbapenem resistance was evaluated for all organisms based on testing resistant to any agent in the carbapenem class. Carbapenem-resistant Enterobacteriaceae (CRE) was based on agent minimum inhibitory concentrations and organism isolated according to definitions by the Clinical and Laboratory Standards Institute.31 For Escherichia sp., Klebsiella sp., Enterobacter sp., Salmonella sp., Shigella sp., Serratia sp. or Citrobacter sp., CRE was defined as intermediate or resistant minimum inhibitory concentrations for the carbapenem class (exception ertapenem=resistant only) and resistant to all tested third-generation cephalosporins. Proteus sp., Providencia sp. and Morganella sp. were similarly defined with the exception of displaying nonsusceptible minimum inhibitory concentrations to at least two of the following: imipenem, meropenem, and/or doripenem.
Statistical analysis
Antimicrobial susceptibility results of individual isolates were compiled into a standardized SCI/D-specific antibiogram and compared to a compiled non-SCI/D antibiogram using Chi-square tests and Fisher’s Exact test as appropriate. Comparison of susceptibilities by location were similarly conducted within each group. Demographics of persons were compared using Student’s t-test or Chi-square. SAS 9.3 (SAS, Inc, Cary, NC,USA) was used for data and statistical analyses. A P-value<0.05 was considered significant.
Results
During the study period, there were a total of 5131 unique cultures for 3135 unique patients. Veterans with SCI/D had 23.0% (n=1179) of unique cultures for 249 unique patients. There was a higher mean number of cultures per patient with SCI/D as compared to the overall hospital population (SCI/D=4.7 cultures/patient, overall=1.4 cultures/patient, P-value<0.0001). Overall, those with SCI/D were younger and more likely to be male than those without SCI/D (Table 1). Compared to those without SCI/D, Veterans with SCI/D had a higher Charlson Index (a measure of severity of illness) but were less likely to have cerebrovascular disease, chronic pulmonary disease, heart failure or renal disease (P⩽0.0423 for all). Although those with SCI/D were more likely to be hospitalized within the past 30 and 90 days (P⩽0.0001), the frequency of surgical procedures within the past 30 and 90 days did not differ between groups (P=NS).
Frequencies of pathogens isolated in the two groups differed. The most frequent pathogens identified in Veterans with SCI/D were Enterococcus sp. (15.2%), Escherichia coli (14.5%), Pseudomonas aeruginosa (12.2%), Enterococcus faecalis (11.0%), Klebsiella pneumoniae (9.3%) and Proteus mirabilis (9.0%) (Table 2). All of these organisms, except for E. coli, were significantly more likely to be isolated in those with SCI/D compared to those without SCI/D. Compared to those without SCI/D, Staphylococcus aureus, coagulase-negative Staphylococcus, Streptococcus agalactiae and viridans Streptococcus were less likely to be isolated in those with SCI/D.
Key antimicrobial agents and their susceptibilities are reported for select organisms in Table 3. Susceptibilities were generally poorer in those with SCI/D. S. aureus isolates in patients with SCI/D were less susceptible to oxacillin, clindamycin, erythromycin and tetracycline than in isolates from non-SCI/D patients. E. faecalis was less susceptible to gentamicin (for synergy) in SCI/D patients, but there were no differences when stratified by vancomycin susceptibility. As compared to non-SCI/D, methicillin-susceptible S. aureus (MSSA)was less susceptible to clindamycin and erythromycin in SCI/D (P⩽0.04 for both). Although SCI/D isolates of methicillin-resistant S. aureus (MRSA) were less susceptible to clindamycin, they were more susceptible to tetracyclines (P⩽0.04 for both). For every antibiotic tested, K. pneumoniae was significantly less susceptible in patients with SCI/D (P<0.05; Table 3). Similarly, E. coli had lower susceptibilities for all agents tested except for carbapenems. P. mirabilis also demonstrated lower susceptibilities in SCI/D for Beta-lactams, quinolones, gentamicin and tobramycin (P<0.05). The P. aeruginosa isolates from SCI/D were less susceptible to cefepime, ciprofloxacin, gentamicin, imipenem and tobramycin as compared to the isolates in those without SCI/D (P⩽0.02 for all).
Over half (57.1%; n=2928) of the unique cultures were associated with the inpatient setting; 27.1% (n=794) were for Veterans with SCI/D. In all, 42.9% (n=2203) of cultures were taken in the outpatient setting; 17.5% (n=385) in Veterans with SCI/D. In both groups, coagulase-negative Staphylococcus, S. aureus, Enterococcus sp., P. aeruginosa, K. pneumoniae, P. mirabilis, Providencia stuartii and Acinetobacter baumannii were more frequently isolated in the inpatient setting (Table 4). As compared to isolates in those with SCI/D in the inpatient setting, non-SCI/D outpatient cultures had higher frequencies of E. coli. Enterobacter cloacae was isolated more frequently in outpatients with SCI/D, whereas this organism was more frequent in the inpatient setting in those without SCI/D.
Differences were identified in the susceptibility of organisms isolated by location (Table 5). Although there was no difference in the frequency of MRSA or methicillin-resistant coagulase-negative Staphylococcus in inpatient and outpatients, there was a greater frequency of vancomycin resistant Enterococci (VRE) isolates cultured in the inpatient setting, although this difference was not significant in those with SCI/D (SCI/D group: inpatient=11.1%, outpatient=5.0%, P=not significant; non-SCI/D group: inpatient=10.1%, outpatient=2.1%, P<0.01). For both groups, inpatient E. coli isolates were significantly less susceptible for aztreonam, cefepime, ceftriaxone, ciprofloxacin and piperacillin/tazobactam (P⩽0.04). For patients without SCI/D, E. coli inpatient isolates were less susceptible to cefazolin, ampicillin/sulbactam, gentamicin and tobramycin (P⩽0.03). Interestingly, there were no common differences in susceptibilities by culture location for K. pneumoniae. For inpatient SCI/D patients, K. pneumoniae isolates, ciprofloxacin, piperacillin/tazobactam, tobramycin and amikacin were significantly less susceptible (P<0.05). In patients without SCI/D, K. pneumoniae was less susceptible to cefazolin and gentamicin. P. aeruginosa isolates cultured in the inpatient setting were less susceptible to piperacillin/tazobactam for both groups (P<0.04). Additionally, non-SCI/D inpatient isolates were less susceptible to cefepime, imipenem and tobramycin for P. aeruginosa (P<0.03).
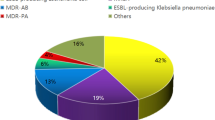
Significantly more ESBL-producing organisms were isolated in patients with SCI/D (P<0.0001). E. coli produced ESBLs most frequently overall and occurred significantly more frequently in the non-SCI/D group (P<0.001; Table 6). Conversely, P. mirabilis and K. pneumoniae were more frequently ESBL-producers in Veterans with SCI/D. Resistance to at least one agent in the carbapenem class occurred in 3.6% of unique isolates. The SCI/D patients had significantly more isolates with carbapenem resistance (7.6% of all SCI/D isolates) as compared to those without SCI/D (2.4% of all non-SCI/D isolates, P<0.0001). Although there were few isolates meeting the definition of CRE (n=21), CREs were more common in the SCI/D group (2.4% vs 0.5%; P<0.0001). Multi-drug resistance occurred in 36.6% of all Gram-negative isolates in the sample: 60.7% of SCI/D and 28.0% of non-SCI/D cultures (P=0.0001). For multi-drug resistance isolates, susceptibilities exceeded 80% only for amikacin, ertapenem and imipenem. With the exception of ampicillin/sulbactam, susceptibilities for multi-drug resistant isolates were generally poorer in Veterans with SCI/D (Table 6).
Discussion
Increasing antimicrobial resistance in complex infections can result in inadequate empiric prescribing. Antibiograms are commonly used tools to guide empiric selection of antimicrobial agents based on bacterial susceptibility patterns at a specific hospital. The lack of rapid and accurate diagnostic tests for pathogen identification and antimicrobial susceptibility is a challenge for all providers, especially for those who care for populations such as SCI/D, with high rates of bacterial resistance. Although the use of an antibiogram to guide empirical antimicrobial treatment has been well documented in the hospital setting,32 the applicability of an antibiogram to SCI/D patients has previously not been explored. As our results demonstrate, not only were the organisms isolated different, but susceptibilities were also generally poorer in Veterans with SCI/D. This difference was especially meaningful in Gram-negative organisms. In addition, inpatient isolates in patients without SCI/D were generally less susceptible where few differences were identified by culture location within the SCI/D group. Patients with SCI/D have multiple risk factors for resistant organisms,23, 24 and our data reflect this increased risk. Carbapenem resistance, MRSA, ESBLs, CREs and multi-drug resistant Gram-negatives were significantly more frequent in Veterans with SCI/D.
Antibiograms are a resource to trend bacterial susceptibility over time, assist with formulary decisions, identify antimicrobial overuse and detect outbreaks and lapse of infection-prevention strategies. Use of local susceptibility data, which is contained in an antibiogram, is also recommended to be used when formulating an antimicrobial stewardship program.15, 32 Although antibiograms are a valuable tool, it has been reported that half of SCI/D providers do not use the facility antibiogram to guide empiric treatment regimens.25 The reasons for this lack of use have not been explored, but it is possible that SCI/D providers are aware that bacterial susceptibility patterns in the overall facility-wide antibiogram do not reflect those observed in patients with SCI/D. Providing a customized antibiogram composed of only SCI/D isolates may improve empiric antibiotic prescribing as the information contained in the antibiogram better reflects those pathogens causing infection.
Comparison of bacterial susceptibilities patterns to develop an SCI-specific antibiogram is not without limitations. The microbiology laboratory reports that were collected and analyzed in this study represent all non-surveillance cultures from all sources obtained from patients regardless of the hospital location. Cultures from different sources may have differing susceptibilities, and consideration of the source of the infection is critical to therapy selection (for example, urine vs blood). Our definition of multi-drug resistance did not account for intrinsic resistance and thus may be an overestimate of high-level resistance for each Gram-negative organism. In addition, the cultures and susceptibilities represented in an antibiogram are highly dependent on the geographic location and the population served. Therefore, our results of bacterial susceptibilities may not apply to the entire SCI/D population. Although the results presented herein are representative of the Veteran population at our facility, they may not be generalizable to other patients seen in other facilities. General limitations of antibiograms include inability to differentiate colonization from infection and lack of patient-specific characteristics and isolate source.
To our knowledge, this is the first report comparing the results of an antibiogram prepared specifically from cultures obtained from patients with SCI/D to cultures obtained from a non-SCI/D population. Although previous studies have described patterns in susceptibility of urinary pathogens in the SCI/D setting,33, 34, 35 the overall prevalence of bacteria responsible for causing disease and the corresponding susceptibilities in this patient population are still largely unknown. In addition, the changing epidemiology of infection and bacterial resistance requires more recent data. In the results presented herein, the frequency of isolation and corresponding susceptibilities of several of the most commonly isolated bacteria from SCI/D patients revealed significant differences compared with the non-SCI/D antibiogram, for which susceptibilities were generally decreased. Thus, reliance on the hospital antibiogram for selecting initial antimicrobial therapy for SCI/D patients could potentially lead to inadequate empiric treatment. It has been found in previous studies that over one-third of patients with SCI/D received inadequate empiric treatment with an antibiotic.36, 37 The availability of more representative data from patients with SCI/D, in the form of an SCI-specific antibiogram, would not only be highly useful in guiding selection of an empiric antimicrobial agent, but may also result in the preservation of antimicrobials through prudent and judicious selection. In addition, the availability of antibiograms specific to certain care areas could offer an avenue for expansion of antimicrobial stewardship efforts customized to the population, including education and improved empirical antimicrobial prescribing.
Different pathogens with poorer susceptibilities are causing infections in patients with SCI/D. However, in a population known to be colonized with multi-drug resistant organisms, future work should be conducted to determine the frequency of colonization vs infection. Thus antimicrobial stewardship is important in this population. Developing stewardship resources, such as unit-specific antibiograms, may be useful in increasing appropriateness of empiric antibiotic treatment.
Data archiving
There were no data to deposit.
References
Costelloe C, Metcalfe C, Lovering A, Mant D, Hay AD . Effect of antibiotic prescribing in primary care on antimicrobial resistance in individual patients: systematic review and meta-analysis. BMJ 2010; 340: c2096.
Ibrahim EH, Sherman G, Ward S, Fraser VJ, Kollef MH . The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 2000; 118: 146–155.
McDonald JR, Friedman ND, Stout JE, Sexton DJ, Kaye KS . Risk factors for ineffective therapy in patients with bloodstream infection. Arch Intern Med 2005; 165: 308–313.
Leibovici L, Konisberger H, Pitlik SD, Samra Z, Drucker M . Patients at risk for inappropriate antibiotic treatment of bacteraemia. J Intern Med 1992; 231: 371–374.
Leibovici L, Paul M, Paznanski O, Drucker M, Samra Z, Konigsberger H et al. Monotherapy versus beta-lactam-aminoglycoside combination treatment for Gram-negative bacteremia: a prospective, observational study. Antimicrob Agents Chemother 1997; 41: 1127–1133.
Leibovici L, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitlik SD . The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J Intern Med 1998; 244: 379–386.
Zaragoza R, Artero A, Camarena JJ, Sancho S, Gonzalex R, Nogueira JM . The influenza of inadequate empirical antimicrobial treatment on patients with bloodstream infections in an intensive care unit. Clin Microbiol Infect 2003; 9: 412–418.
Chaubey VP, Pitout JD, Dalton B, Ross T, Church DL, Gregson DB et al. Clinical outcomes of empiric antimicrobial therapy of bacteremia due to extended-spectrum beta-lactamase producing Escherichia coli and Klebsiella pneumonia. BMC Res Notes 2010; 3: 116.
Kang CI, Kim SH, Kim HB, Park SW, Choe YJ, Oh MD et al. Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin Infect Dis 2003; 37: 745–751.
Kang CI, Kim SH, Park WB, Lee KD, Kim HB, Kim EC et al. Bloodstream infections caused by antibiotic-resistant Gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother 2005; 49: 760–766.
Harbarth S, Garbino J, Pugin J, Romand JA, Lew D, Pittet D . Inappropriate initial antimicrobial therapy and its effect on survival in a clinical trial of immunomodulating therapy for severe sepsis. Am J Med 2003; 115: 529–535.
Micek ST, Lloyd AE, Ritchie DJ, Reichley RM, Fraser VJ, Kollef MH . Pseudomonas aeruginosa bloodstream infection: importance of appropriate initial antimicrobial treatment. Antimicrob Agents Chemother 2005; 49: 1306–1311.
Tumbarello M, Spanu T, Bidino RD, Marchetti M, Ruggeri M, Trecarichi EM et al. Costs of bloodstream infections caused by Escherichia coli and influence of extended-spectrum-beta-lactamases production and inadequate initial antibiotic therapy. Antimicrob Agents Chemother 2010; 54: 4085–4091.
Imamura T, Ohta B, Tanaka E, Branch J . Prognosis of urosepsis patients who are treated by inappropriate initial antimicrobial therapy in the emergency department. Ann Emerg Med 2009; 54: S38–S39.
Xu R, Polk RE, Stencel L, Lowe DK, Guharoy R, Duggal RW et al. Antibiogram compliance in University HealthSystem Consortium participating hospitals with Clinical and Laboratory Standards Institute guidelines. Am J Health Syst Pharm 2012; 69: 598–606.
Fridkin SK, Edwards JR, Tenover FC, Gaynes RP, McGowan JE Jr for the ICARE Project and NNIS. Antimicrobial resistance prevalence rates in hospital antibiograms reflect prevalence rates among pathogens associated with hospital-acquired infections. Clin Infect Dis 2001; 33: 324–329.
Kaufman D, Haas CE, Edinger R, Hollick G . Antibiotic susceptibility in the surgical intensive care unit compared with the hospital-wide antibiogram. Arch Surg 1998; 133: 1041–1045.
Draper HM, Farland JB, Heidel RE, May LS, Suda KJ . Comparison of bacteria isolated from emergency department patients versus hospitalized patients. Am J Health Syst Pharm 2013; 70: 2124–2128.
Healthcare Analysis and Information Group (HAIG) Office of the Assistant Deputy Under Secretary for Health for Policy and Planning. 2012 Survey of Antimicrobial Stewardship in VHA. Department of Veterans Affairs, Washington, DC, USA, May 2013.
Binkley S, Fisherman NO, LaRosa LA, Marr AM, Nachamkin I, Wordell D et al. Comparison of unit-specific and hospital-wide antibiograms potential implications for selection of empirical antimicrobial therapy. Infect Control Hosp Epidemiol 2006; 27: 682–687.
Pogue JM, Alaniz C, Carver PL, Pleva M, Newton D, DePestel DD . Role of unit-specific combination antibiograms for improving the selection of appropriate empiric therapy for Gram-negative pneumonia. Infect Control Hosp Epidemiol 2011; 32: 289–292.
Swami SK, Banerjee R . Comparison of hospital-wide and age and location-stratified antibiograms of S. aureus E. coli, and S. pneumoniae: age-and location-stratified antibiograms. SpringerPlus 2013; 2: 63.
Montgomerie JZ . Infections in patients with spinal cord injuries. Clin Infect Dis 1997; 25: 1285–1290.
Evans CT, LaVela SL, Weaver FM, Priebe M, Sandford P, Niemiec P et al. Epidemiology of hospital-acquired infections in Veterans with spinal cord injury and disorder. Infect Control Hosp Epidemiol 2008; 29: 234–242.
Evans CT, Rogers TJ, Burns SP, Lopansri B, Weaver FM . Knowledge and use of antimicrobial stewardship resources by spinal cord injury providers. PM R 2011; 3: 619–623.
Goff DA . Antimicrobial stewardship: bridging the gap between quality care and cost. Curr Opin Infect Dis 2011; 24: S11–S20.
Hindler JF, Stelling J . Analysis and presentation of cumulative antibiograms: a new consensus guideline from the Clinical and Laboratory Standards Institute. Clin Infect Dis 2007; 44: 867–873.
Smith B, Evans CT, Ullrich P, Burns S, Guihan M, Miskevics S et al. Using VA data for research in persons with spinal cord injuries and disorders: lessons from SCI QUERI. J Rehabil Res Dev 2010; 47: 679–688.
CLSI. Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data, 3rd edn, Guideline M39-A3. Clinical and Laboratory Standards Institute: Wayn, PA, USA, 2009.
Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing; Twenty-third Informational Supplement M100-S23. CLSI: Wayne, PA, USA. 2013.
Clinical and Laboratory Standards Institute Interpretive Criteria for Carbapenems and Enterobacteriaceae M100-S22. CLSI: Wayne, PA, USA. 2012.
Dellit TH, Owens RC, McGowan JE Jr, Gerding DN, Weinstein RA, Burke JP et al. Infectious Diseases Society of America; Society for Healthcare Epidemiology of America. Infectious Diseases Society of America and Society for Healthcare Epidemioloogy of America guidelines for developing an institutional program to enhance antimicrobial stewardship. Clin Infect Dis 2007; 44: 159–177.
Yoon SB, Lee BS, Lee KD, Hwang SI, Lee HJ, Han ZA . Comparison of bacterial strains and antibiotic susceptibilities in urinary isolates of spinal cord injury patients from the community and hospital. Spinal Cord 2014; 52: 298–301.
Waites KB, Chen Y, DeVivo MJ, Canupp KC, Moser SA . Antimicrobial resistance in Gram-negative bacteria isolated from the urinary tract in community-residing person with spinal cord injury. Arch Phys Med Rehabil 2000; 81: 764–769.
Hinkel A, Finke W, Botel U, Gatermann SG, Pannek J . Increasing resistance against antibiotics in bacteria isolated from the lower urinary tract of an outpatient population of spinal cord injury patients. Urol Int 2004; 73: 143–148.
Montgomerie JAZ, Chan E, Gilmore DS, Canawati HN, Sapico FL . Low mortality among patients with spinal cord injury and bacteremia. Rev Infect Dis 1991; 13: 867–871.
Evans CT, Burns SP, Chin A, Weaver FM, Hershow RC . Predictors and outcomes of antibiotic adequacy for bloodstream infections in Veterans with spinal cord injury. Arch Phys Med Rehabil 2009; 90: 1364–1370.
Acknowledgements
The views expressed in this article are those of the authors and do not necessarily represent the views of the Department of Veterans Affairs, the Health Services Research and Development Service or the United States government. This work was supported by the US Department of Veterans Affairs as a QUERI center grant (SCI 98-001) and QUERI LIP (QLP 97-009).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This project was presented, in part, as a poster at the Infectious Disease Society of America Annual Meeting in Philadelphia, PA, USA, October 2014.
Rights and permissions
About this article
Cite this article
Suda, K., Patel, U., Sabzwari, R. et al. Bacterial susceptibility patterns in patients with spinal cord injury and disorder (SCI/D): an opportunity for customized stewardship tools. Spinal Cord 54, 1001–1009 (2016). https://doi.org/10.1038/sc.2016.38
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sc.2016.38
This article is cited by
-
Identifying prescribers of antibiotics in a primary care spinal cord injury cohort
Spinal Cord Series and Cases (2024)
-
The prevalence of antibiotic-resistant and multidrug-resistant bacteria in urine cultures from inpatients with spinal cord injuries and disorders: an 8-year, single-center study
BMC Infectious Diseases (2022)
-
Epidemiology and outcomes associated with carbapenem-resistant Acinetobacter baumannii and carbapenem-resistant Pseudomonas aeruginosa: a retrospective cohort study
BMC Infectious Diseases (2022)
-
Multidrug-resistant bacteria in urine culture among patients with spinal cord injury and disorder: Time to first detection and analysis of risk factors
Spinal Cord (2022)
-
Effect of probiotics on multi-resistant organism colonisation in persons with spinal cord injury: secondary outcome of ProSCIUTTU, a randomised placebo-controlled trial
Spinal Cord (2020)