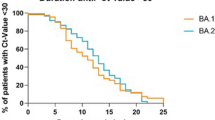
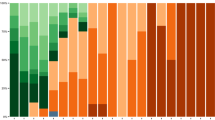
Abstract
Background
The genetic and pathogenic characteristics of SARS-CoV-2 have evolved from the original isolated strains; however, the changes in viral virulence have not been fully defined. In this study, we analyzed the association between the severity of the pathogenesis of pneumonia in humans and SARS-CoV-2 variants that have been prevalent to date.
Methods
We examined changes in the variants and tropism of SARS-CoV-2. A total of 514 patients admitted between February 2020 and August 2022 were included and evaluated for pneumonia by computed tomography (CT) as a surrogate of viral tropism.
Results
The prevalence of pneumonia for each variant was as follows: D614G (57%, 65/114), Alpha (67%, 41/61), Delta (49%, 41/84), Omicron BA.1.1 (26%, 43/163), and Omicron BA.2 (11%, 10/92). The pneumonia prevalence in unvaccinated patients progressively declined from 70% to 11% as the variants changed: D614G (56%, 61/108), Alpha (70%, 26/37), Delta (60%, 38/63), BA.1.1 (52%, 15/29), and BA.2 (11%, 2/19). The presence of pneumonia in vaccinated patients was as follows: Delta (16%, 3/19), BA.1.1 (21%, 27/129), and BA.2 (11%, 8/73). Compared with D614G, the areas of lung involvement were also significantly reduced in BA.1.1 and BA.2 variants.
Conclusions
Compared with previous variants, there was a marked decrease in pneumonia prevalence and lung involvement in patients infected with Omicron owing to decreased tropism in the lungs that hindered viral proliferation in the alveolar epithelial tissue. Nevertheless, older, high-risk patients with comorbidities who are infected with an Omicron variant can still develop pneumonia and require early treatment.
Plain language summary
The SARS-CoV-2 virus changes over time with the differing viruses described as variants. The different variants of SARS-CoV-2 have an impact on how easily they infect people and the effects they have on infected individuals. Here, we examined images of the lungs of patients hospitalized with COVID-19 to investigate whether they had pneumonia, a type of swelling in the lung. Compared with the variant found early in the pandemic, the more recent Omicron variant led to a decreased rate of pneumonia in infected individuals. Our findings emphasize the need for early treatment, as pneumonia may progress in older patients or those with other illnesses.
Similar content being viewed by others
Introduction
Since its emergence in 2019, SARS-CoV-2 has caused an enormous number of infections and deaths. However, the population is becoming increasingly immune owing to an increase in the number of natural infections and increasing numbers of vaccinated individuals. In some areas, mass immunity has been achieved, and infection mitigation is gradually progressing1,2,3. In addition, therapeutics such as monoclonal antibodies, antiviral drugs, and anti-inflammatory drugs have been developed that have helped to reduce the risk of infection and the severity of COVID-194. The virus has accumulated mutations over time5, and the emergence of more infectious viral variants has led to a temporary increase in infections, but a decrease in viral virulence has also occurred6. Breakthrough infections can occur in individuals who have received more than two doses of vaccine7,8,9. Against this background, to improve infection control, we must clearly understand the infectivity and virulence of SARS-CoV-2 by analyzing the data that have accumulated over the past 2.7 years.
In Japan, national guidelines recommend admitting COVID-19 patients to the hospital even if their symptoms are mild enough to allow them to stay at home. Japan has a comprehensive national health insurance system that allows for chest computed tomography (CT) scans even in asymptomatic patients. Chest CT scanning can detect subtle pulmonary lesion changes, such as the peripherally distributed bilateral and multilobar ground-glass opacities, known as typical image of COVID-19 pneumonia. We previously performed whole-genome sequencing to determine genotype changes in SARS-CoV-2 over 2.7 years10,11,12,13. Data regarding the presence or absence of pneumonia in a wide range of clinical conditions have also accumulated.
In the current study, we investigated changes in lung viral tropism in COVID-19 patients by examining changes in the SARS-CoV-2 variants and the prevalence of pneumonia in vaccinated and unvaccinated patients. Results showed a higher risk of pneumonia with older age, male gender, and the Alpha variant, while being fully vaccinated and having the Omicron BA.2 variant were associated with a lower risk. The study also found differences in disease severity and outcomes between variants. These findings have important clinical implications for the management of COVID-19 and may inform vaccination and treatment decisions.
Methods
Patients
This study included 730 hospitalized patients who tested positive for SARS-CoV-2 infection by PCR and/or antigen assay between February 2020 and August 2022. We excluded 84 hospitalized patients who did not undergo SARS-CoV-2 genomic analysis due to low viral loads. We also excluded 21 individuals who initially tested positive for SARS-CoV-2 by PCR but were likely false positives according to subsequent analyses14. A further 110 patients, including children or pregnant women who had not undergone CT scanning, were also excluded. Only one patient with the Gamma variant was detected during this period, but this case was removed from subsequent analyses because of the small number of Gamma variant detections13.
In total, 514 hospitalized patients were finally included in this study. The CT images of the lungs were independently evaluated for the presence of pneumonia by a radiologist and two respiratory physicians. COVID-19-typical pneumonia was identified as peripherally distributed bilateral and multilobar ground-glass opacities, which are characteristic CT findings that have been reported in COVID-19 pneumonia15,16. Medical staff collected data on the medical condition, vaccination status, and disease course from patient interviews and retrospectively examined the electronic medical records. People who had received at least two vaccine doses with more than 14 days passing since the second dose were considered fully vaccinated. Breakthrough infection was defined as a positive SARS-CoV-2 PCR or antigen test result in a fully vaccinated individual.
The Institutional Review Board of the Clinical Research and Genome Research Committee at our hospital approved this study and the use of an opt-out consent method (approval no. C2019-30). The requirement for written informed consent was waived because this was an observational study, and there is an urgent need to collect COVID-19 data.
SARS-CoV-2 diagnostic testing
Multiple molecular diagnostic platforms of nucleic acid amplification and antigen testing were used to diagnose SARS-CoV-2 infection. The diagnostic tests used were reverse transcription PCR (in accordance with the protocol developed by the National Institute of Infectious Diseases in Japan17), the FilmArray Respiratory Panel 2.1 test with the FilmArray Torch system (bioMérieux, Marcy-l’Etoile, France)18, the Xpert Xpress SARS-CoV-2 test using Cepheid GeneXpert (Cepheid, Sunnyvale, CA)19, and the Lumipulse antigen test with the LUMIPULSE G600II system (Fujirebio, Inc., Tokyo, Japan)20,21. All tests were performed with material obtained from nasopharyngeal swabs immersed in viral transport medium (Copan, Murrieta, CA).
SARS-CoV-2 whole-genome analysis
Whole-genome sequencing analysis was conducted on SARS-CoV-2-positive samples collected from hospitalized patients as previously described11. In brief, SARS-CoV-2 genomic RNA was reverse-transcribed into cDNA and amplified using the Ion AmpliSeq SARS-CoV-2 Research Panel or the Ion AmpliSeq SARS-CoV-2 Insight Research Assay (Thermo Fisher Scientific, Waltham, MA) on the Ion Torrent Genexus system according to the manufacturer’s instructions11,13. Sequencing reads were processed and quality was assessed using Genexus software with SARS-CoV-2 plugins. The sequencing reads were then mapped and aligned using the Torrent Mapping Alignment Program. After initial mapping, a variant call was performed using Torrent Variant Caller. The COVID19AnnotateSnpEff plugin was used to annotate the variants. Assembly was performed using Iterative Refinement Meta-Assembler22.
The viral clade and lineage classifications were conducted using Nextstrain23 and Phylogenetic Assignment of Named Global Outbreak Lineages24. Sequence data were deposited in the Global Initiative on Sharing Avian Influenza Data EpiCoV database5.
CT imaging
All patients underwent CT immediately after arriving at the hospital. CT images (64 × 0.5 mm or 32 × 1.0 mm-detector row/automatic exposure control/120 kVp) were obtained with the Aquilion 64 or the Aquilion 64CX system (Canon Medical, Tochigi, Japan). The 0.5 mm or 1.0 mm-slice thickness high-resolution computed tomography images were reconstructed using the FC51 reconstruct function.
CT image analysis
All CT images were evaluated by an experienced radiologist specialized in thoracic radiology with 25 years of experience. Definitions of radiological terms are based on the standard glossary of terms for chest imaging reported by the Fleischner Society25. On the basis of previous reports, a pneumonia diagnosis was made and the main findings on chest CT were classified into three categories: consolidation (CON), crazy-paving appearance (CPA), and ground-glass opacity (GGO)26,27,28. The distribution of lung abnormalities was divided into three sections from the pulmonary hilum to the subpleural area and classified as those that were only subpleural (peripheral), those that extended near to the pulmonary hilum (central), and those that extended to the middle area between the peripheral and central areas. Semi-quantitative CT scores were evaluated for the right upper lobe, right middle lobe, right lower lobe, left upper lobe, and left lower lobe based on previous reports29. Briefly, the CT scores were calculated according to the lesion area as follows: 0, 0% involvement; 1, <5% involvement; 2, 5%–25% involvement; 3, 26%–50% involvement; 4, 50%–75% involvement; 5, >75% involvement. The total CT score was the sum of the individual scores, ranging from 0 (no involvement) to 25 (maximum involvement).
Serological analysis
We measured the titers of anti-nucleocapsid (N) protein antibody and anti-spike (S) protein receptor-binding domain antibody using the Elecsys Anti-SARS-CoV-2 antibody test (Roche Diagnostics, Basel, Switzerland) on a cobas® 8000 automated platform14. This assay utilizes the electrochemiluminescence immunoassay principle. For the anti-N antibody, samples with a cutoff index (COI) < 1.0 were considered negative, while those with a COI ≥ 1.0 were considered positive. For the anti-S antibody, samples containing <0.8 U/mL were considered negative, while those containing ≥0.8 U/mL were considered positive, in accordance with the manufacturer’s instructions.
Statistical analysis
All statistical tests and visualizations were performed with R, version 4.1.1 (R Foundation for Statistical Computing) or RStudio (https://www.rstudio.com/). The following R packages were used for data cleaning, analysis, and visualization: ggplot2 (v3.3.5), dplyr (v1.0.7), tidyr (v1.1.3), patchwork (v1.1.1), gtsummary (v1.5.2), and flextable (v.0.7.0). A multivariate logistic regression analysis was performed to evaluate the effects of age (over or under 65 years), sex, viral lineage, and vaccination status on the likelihood of having COVID-19 pneumonia. Odds ratios (ORs) and the corresponding 95% confidence intervals (CIs) were calculated. P-values <0.05 were considered to be statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Pneumonia prevalence and clinical background
Patients admitted to our hospital between February 2020 and August 2022 were included in the study. Of the 730 hospitalized patients, 514 were evaluated for pneumonia by CT scanning, and patient samples were sequenced to identify the SARS-CoV-2 variant. Of these 514 patients (median age: 66.5 years [interquartile range (IQR), 46–81], female: 219, male: 295), 314 (61%) had no COVID-19-typical pneumonia, while 200 (39%) had COVID-19-typical pneumonia (Supplementary Data 1). Compared with patients without pneumonia, those with pneumonia tended to be male (P = 8.6 × 10−4), unvaccinated (P = 8.0 × 10−18), require supplemental oxygen (P = 8.1 × 10−76) and ventilator support (P = 0.016), and have worse outcomes (P = 0.002) (Supplementary Data 1). Compared with D614G, Alpha, and Delta variants, patients infected with Omicron BA.1.1 or BA.2 variants had significantly lower rates of pneumonia (D614G vs. BA.1.1, adjusted P = 2.6 × 10−6 D614G vs. BA.2, adjusted P = 1.9 × 10−11 Alpha vs. BA.1.1, adjusted P = 2.3 × 10−7 Alpha vs. BA.2, adjusted P = 4.9 × 10−12 Delta vs. BA.1.1, adjusted P = 3.1 × 10−3 Delta vs. BA.2, adjusted P = 1.8 × 10−7, pairwise Fisher’s exact test) (Supplementary Table 1). Of the 38 breakthrough infections with pneumonia, remdesivir was administered in 29 (76%), casirivimab/imdevimab in 1 (2.6%), sotrovimab in 2 (5.3%), and molnupiravir in 4 (11%) cases; nirmatrelvir/ritonavir were not used for treatment (Supplementary Table 2).
Multivariable logistic regression analysis
A multivariate logistic regression model was applied to examine independent predictors contributing to the presence or absence of COVID-19-typical pneumonia as determined by CT and their corresponding ORs and 95% CIs. The following factors were associated with a higher risk of COVID-19 pneumonia: 65 years and older (OR = 3.38; 95% CI, 2.02–5.83), being male (OR = 2.14; 95% CI, 1.38–3.36), and the Alpha variant (OR = 2.39; 95% CI, 1.05–5.71) (Table 1). The BA.2 variant (OR = 0.20; 95% CI, 0.08–0.46) and being fully vaccinated (OR = 0.17; 95% CI, 0.09–0.33) were associated with a lower risk of developing COVID-19 pneumonia (Table 1).
Variants and CT pneumonia images
The number of patients hospitalized with COVID-19 has intermittently increased over the past 2.7 years (Fig. 1a). The variants identified by whole-genome sequencing during the observation period were D614G (n = 114), Alpha (n = 61), Delta (n = 84), Omicron BA.1.1 (n = 163), and Omicron BA.2 (n = 92).
a Historical transition of SARS-CoV-2 variants identified from February 2020 to August 2022. During this observation period, we identified the following numbers of hospitalized patients infected with the variants: D614G (n = 114), Alpha (n = 61), Delta (n = 84), BA.1.1 (n = 163), and BA.2 (n = 92). b Number of patients (left panel) and percentage of patients (right panel) that developed pneumonia by variant. c Number (left panel) and percentage (right panel) of patients requiring supplemental oxygen by variant. d Number (left panel) and percentage (right panel) of patients requiring ventilator use by variant. e Number (left panel) and percentage (right panel) of patient outcomes by variant. Numbers above bars indicate number of cases.
The CT findings of COVID-19-typical pneumonia were examined in detail. Pneumonia was observed in patients infected with the D614G (57%, 65/114), Alpha (67%, 41/61), Delta (49%, 41/84), Omicron BA.1.1 (26%, 43/163), and Omicron BA.2 (11%, 10/92) variants (Fig. 1b). There was a substantial decrease in the presence of pneumonia between the Alpha (67%) and Delta (49%) variants compared with the Omicron BA.1.1 (26%) and Omicron BA.2 (11%) variants (Fig. 1b).
Variant, supplemental oxygen and ventilator use, and outcomes
If a patient’s arterial oxygen saturation decreased, the use of supplemental oxygen was initiated regardless of the cause. Supplemental oxygen was required for patients infected with the D614G (65%, 74/114), Alpha (72%, 44/61), Delta (49%, 41/84), Omicron BA.1.1 (39%, 63/163), and Omicron BA.2 (20%, 18/92) variants (Fig. 1c). The number of patients requiring supplemental oxygen decreased as the viral genotype successively changed. For those infected with Omicron BA.2, 20% of patients required supplemental oxygen, which was higher than the 11% of patients with pneumonia identified by chest CT. However, for eight of these patients, oxygen was supplied for other reasons (e.g., aspiration pneumonia) and was not considered to be a result of COVID-19 severity.
The number of patients requiring ventilator use also successively reduced as the variants changed: D614G (6%, 7/114), Alpha (2%, 1/61), Delta (0%, 0/84), Omicron BA.1.1 (1%, 1/163), and Omicron BA.2 (1%, 1/92) (Fig. 1d). As the virus evolved, the number of patients requiring ventilator use approached zero. The following numbers of patients died after hospital admission: D614G (5%, 6/114), Alpha (2%, 1/61), Delta (0%, 0/84), Omicron BA.1.1 (4%, 7/163), and Omicron BA.2 (1%, 1/92) (Fig. 1e).
SARS-CoV-2 variant according to vaccination status and pneumonia findings
We examined the variant according to vaccination status for the 477 individuals with a known vaccination history; this analysis excluded the 37 individuals with an unknown vaccination history. We observed the following numbers of variant infections among those who were fully vaccinated (i.e., those who had received at least two vaccine doses): D614G (0%, 0/108), Alpha (0%, 0/37), Delta (23%, 19/82), BA.1.1 (82%, 129/158), and BA.2 (79%, 73/92) (Fig. 2a). The number of people who were fully vaccinated increased sharply from 0% for those infected with D614G to 82% for those infected with BA.1.1 but was relatively stable at 79% for those infected with BA.2 (Fig. 2a).
a Vaccination status by variant. Number of patients (left panel) and percentage of patients (right panel) classified as unvaccinated or fully vaccinated (i.e., two or more doses). b, c Presence or absence of pneumonia by variant in unvaccinated (b) and fully vaccinated patients (c). Numbers above bars indicate number of cases.
Next, we examined the variant according to pneumonia findings and vaccination status. The number of unvaccinated patients with COVID-19-typical pneumonia remained almost unchanged across the variants (D614G (56%, 61/108), Alpha (70%, 26/37), Delta (60%, 38/63), and BA.1.1 (52%, 15/29) (Fig. 2b). However, for those infected with the BA.2 genotype, fewer unvaccinated patients displayed pneumonia symptoms (11%, 2/19) (Fig. 2b).
We also examined the presence of pneumonia in fully vaccinated patients. None of the patients infected with either D614G or Alpha were fully vaccinated; therefore, these patients were not assessed further. COVID-19-typical pneumonia was apparent in 16% (3/19) and 21% (27/129) of vaccinated patients infected with Delta and BA.1.1, respectively; this is a 1/3 to 1/4 decrease compared with unvaccinated patients (60% and 52%) (Fig. 2c). In contrast, vaccinated and unvaccinated BA.2-infected patients showed similar rates of COVID-19-typical pneumonia by CT imaging (vaccinated: 11%, 8/73; unvaccinated: 11%, 2/19) (Fig. 2c).
SARS-CoV-2 variants and lung involvement
A quantitative CT score was calculated to evaluate the changes in pulmonary involvement in patients infected with the different variants29. The median total CT score in patients infected with D614G was 5 (IQR: 0.25–9), Alpha was 10 (IQR: 2–14), Delta was 2 (IQR: 0–11), BA.1.1 was 2 (IQR: 0–7), and BA.2 was 1 (IQR: 0–4) (Fig. 3). Compared with D614G, the total CT scores of patients with Alpha were significantly higher (P = 0.031), whereas those with BA.1.1 (P = 0.031) and BA.2 (P = 7.0 × 10–5) were significantly lower (Fig. 3, P-values were adjusted for multiple comparisons). Except for the right middle lobe, the CT scores showed similar trends in the other lung lobes according to the SARS-CoV-2 variant (Supplementary Fig. 1). These results suggest that an Omicron infection does not spread as readily into the lungs.
The CT scores were evaluated semi-quantitatively based on the percentage of lung involvement in the five lung lobes, and the sum of the CT scores (total CT score) was calculated. Box plots show the total CT scores of D614G (n = 114), Alpha (n = 61), Delta (n = 84), BA.1.1 (n = 163), and BA.2. (n = 92). Each box indicates the interquartile range (top: the third quartile; bottom: the first quartile) with a horizontal line indicating the median. Statistical analysis was performed by a t-test, and P-values were adjusted for multiple comparisons.
Breakthrough infection in vaccinated BA.2-infected patients with pneumonia
We next analyzed the BA.2-infected breakthrough infection cases in detail. Eight BA.2-infected patients developed COVID-19-typical pneumonia according to the CT images (Fig. 4 and Supplementary Data 2). Case #8 showed atypical consolidation because the left lower lung was atelectatic. However, based on the clinical course of the disease, we diagnosed the patient with COVID-19 typical pneumonia because the right lung showed ground-glass shadows. The median age of these patients was 79 years (range 68–90 years), with three females and five males. Comorbidities included interstitial pneumonia (n = 1), hyperlipidemia (n = 1), diabetes (n = 2), hypertension (n = 2), and cancer (n = 1) (Supplementary Data 2). There were no secondary complications such as lung embolism, pneumothorax, or pneumoperitoneum. Of these patients, seven survived and one passed away.
One patient had received two vaccine doses, and seven had received three vaccine doses. All patients were vaccinated with BNT162b2 (Supplementary Data 2). The median duration from the time of last vaccination to infection was 6.6 weeks (range: 2.4–26.7 weeks). The anti-N antibody titers were negative in all cases, with a COI range of 0.07–0.17, implying that these patients had no previous history of infection. The pneumonia distributions were central (n = 2), peripheral (n = 1), and peripheral and central (n = 5), and the main patterns were CON (n = 2), CPA (n = 2), and GGO (n = 4).
We investigated whether the time between the most recent vaccination and infection was associated with disease progression. The anti-S antibody titers decreased with time since the most recent vaccination (R = ˗0.94, P = 4.6 × 10−4) (Supplementary Fig. 2a), but no correlation was found between this time period and the CT score (R = 0.23, P = 0.59) (Supplementary Fig. 2b). In addition, although the number of cases was small, the time between vaccination and infection was not associated with the degree of symptoms or the outcome (Supplementary Fig. 2c and 2d, Mann–Whitney U-test).
Pneumonia findings in unvaccinated BA.2-infected patients
The median age of the 19 unvaccinated BA.2-infected patients was 76 years (range 8–92 years), with 12 females and 7 males (Fig. 5). Four of these patients required supplemental oxygen. One of these patients required the use of a ventilator, and all of these patients survived. The median anti-S antibody titer was 0.4 U/mL (range 0.4–9476 U/mL, n = 16). The anti-N antibody titers ranged from 0.07 to 88.5 COI (n = 17). Five patients (cases #15, #17, #20, #21, and #26) had elevated S antibody titers. Case #15, #20, #21, and #26 were considered to have an immune response because sufficient time had passed since the infection. Case #17 had been previously infected with SARS-CoV-2. Of these 19 patients, 10 had no signs of pneumonia, seven showed COVID-19-atypical pneumonia, and only two showed COVID-19-typical pneumonia (Fig. 5 and Supplementary Data 2).
Comparison of unvaccinated and fully vaccinated BA.2-infected patients who had COVID-19-typical pneumonia
In this study, of the ten BA.2-infected patients with COVID-19-typical pneumonia, two were unvaccinated (11%, 2/19) and eight were fully vaccinated (11%, 8/73). Although the vaccination rate was 79% among those infected with BA.2 (Fig. 2a), the number of patients with findings of COVID-19 typical pneumonia remained high, and one patient died. One unvaccinated patient was treated with baricitinib, but not with antiviral or antibody therapy (Supplementary Data 2). Of the eight fully vaccinated patients, five were treated with remdesivir, one with remdesivir + baricitinib + dexamethasone, one with remdesivir + dexamethasone, and one with remdesivir + sotrovimab (Supplementary Data 2). Although not significant, the median total CT score tended to be lower in fully vaccinated patients compared with unvaccinated patients (P = 0.066, Mann–Whitney U-test) (Supplementary Fig. 3). These results suggest that while the vaccine may be effective against the spread of lung lesions, frail and elderly individuals with some comorbidities infected with BA.2 are at risk of developing pneumonia and mortality30, suggesting limitations of vaccines designed based on the original strain.
Discussion
In this study, hospitalized patients underwent CT imaging to assess their pneumonia status. Consecutive data acquisition from the early to the most recent SARS-CoV-2 variant outbreaks showed that a higher proportion of hospitalized patients with pneumonia were male, older, had poorer outcomes, and were unvaccinated. Furthermore, the proportion of hospitalized patients with pneumonia showed a sharp downward trend with the successive changes in SARS-CoV-2 variant. The percentage of hospitalized patients with COVID-19-typical pneumonia peaked at 67% for the Alpha variant and then decreased markedly to 49% for Delta, 26% for BA.1.1, and 11% for BA.2, marking a major decline in pneumonia prevalence from Delta to Omicron. In particular, there was a prominent decrease in pneumonia cases for those infected with BA.2.
Studies in animal models have reported that compared with Delta, the Omicron variants are more infectious in the upper respiratory tract but are less likely to invade the lungs and are less virulent31,32. Consistent with these trends, our results showed a lower rate of pneumonia in hospitalized patients infected with the Omicron variants than with the other SARS-CoV-2 variants (Supplementary Data 1). CT findings showed an organizing pneumonia pattern was typical in the early phase of the pandemic, but a bronchiolitis pattern was more common after the emergence of Omicron. Both the incidence of pneumonia and CT score were lower in Omicron-infected patients, suggesting Omicron is less aggressiveness. This implies a potential scenario in which viral evolution has led to increasing infectivity but decreasing virulence33,34. Nevertheless, old age was identified as a risk factor for developing pneumonia in patients infected with the Omicron after receiving an inactivated vaccine injection35. This data is supported in the present study, which shows that older, high-risk patients with comorbidities who are infected with the Omicron BA.2 variants are still at risk of developing pneumonia and need early treatment to avoid severe disease.
When considering changes in the virulence of SARS-CoV-2, it is crucial to determine whether (1) tropism has changed and infection of the lung tissue has decreased, (2) the changes are due to the effect of vaccination, and (3) whether both of these factors are involved. In particular, compared with the previous variants, the Omicron variants contain more mutations, are more infectious, and have greater immune evasion36. Vaccine effectiveness increases with the third dose37, but the overall vaccine effectiveness has been attenuated over time as the virus has evolved from the Delta variants to the Omicron variants38. Notably, of the eight patients who had a breakthrough infection with BA.2, six developed COVID-19-typical pneumonia despite having a high anti-S antibody titer and a short duration since their third vaccine dose (2.4–10.9 weeks). The anti-S antibody titers may have been raised by the booster dose, but the extent to which these antibodies have neutralizing activity in hospitalized patients needs to be evaluated. The first developed vaccines were designed against the original virus and were highly effective in the early phase of pandemics. However, it should be noted that conventional vaccines may not adequately protect against infection or prevent severe disease because Omicron has numerous mutations in the S gene.
Recent studies have shown that the Omicron variants have reduced virulence resulting from alterations in tropism. The Omicron variants have been reported to utilize cathepsin-dependent endosomal entry more than TMPRSS2-dependent plasma membrane entry32,39,40. Furthermore, Omicron replicates faster in the bronchi than the other SARS-CoV-2 variants, but it is less efficient at infecting the lung parenchyma41. Experimental infection in hamsters showed that BA.2 is more likely than BA.1 to infect lung tissue42, but this was not supported by our clinical data. In the present study, the symptoms of the BA.2-infected patients were due to laryngitis and bronchiolitis, and the patients with BA.2 infection rarely showed clinical conditions consistent with pulmonary parenchymal damage. We speculate that BA.2 has decreased tropism for infection of the alveolar epithelium and may be more confined to the bronchiolar epithelium. Our study showed that lung involvement was significantly less common in BA.2-infected patients, which was in line with the CT scoring evaluation results. In other words, owing to the change in tropism, the pathogenicity of COVID-19 has changed so that it is less likely to result in lung parenchymal damage.
This study had several limitations. First, this dataset included hospitalized patients from only a single institution. Second, there were many confounding factors such as age, vaccination status, the time between the most recent vaccination and infection, and public health measures that complicated examining the relationship between the variants and the presence or absence of pneumonia. Third, although CT scans were performed immediately after admission, they were not performed subsequently; thus, the lung images could not be evaluated over time. Therefore, to accurately assess when COVID-19 pneumonia first appeared and when the condition worsened was not possible. Fourth, there was insufficient information on the history of prior infections for all patients. Ideally, separating the truly immunologically naïve patients from the previously infected patients who had infection-elicited immunity in the unvaccinated group would be preferable. To obtain some information about this, we evaluated the seropositivity of anti-N antibodies as a surrogate indicator of previous infection in the unvaccinated patients. Of the 256 unvaccinated patients, 48 (18.8%) were seropositive for N antibodies at admission, 205 (80.1%) were seronegative, and 3 (1.2%) were untested. Thus, it can be inferred that almost of the unvaccinated patients in our cohort had no history of previous infection.
In summary, the analysis of this dataset obtained over time showed that the proportion of COVID-19 patients with pneumonia decreased as the SARS-CoV-2 variant changed. Using CT images as a surrogate indicator of viral proliferation in the lung parenchyma, and hence pneumonia induction, the results suggest that BA.2 tropism in the lungs is decreased, resulting in a shift to upper respiratory tract proliferation rather than a pneumonia-inducing type of infection. Studies have suggested that governments gradually lessen the rigor of infection control measures and the use of non-pharmaceutical interventions when viral virulence decreases and herd immunity is achieved owing to an increased number of vaccinated individuals and previously infected individuals in the general population43,44. Over time after vaccination, serum antibody levels and vaccine effectiveness decrease, so additional vaccinations may be necessary depending on the disease severity caused by the variant circulating in the community45. In the future, additional vaccination may be promoted preferentially in the older adult population and for those at high risk of disease progression owing to underlying diseases46,47. To be taken into consideration is the fact that the vaccines designed for original virus are already not compatible with the currently circulating Omicron variants because of a poor antigenic match. In that context, the Omicron BA.4/BA.5-adapted bivalent COVID-19 vaccines are expected to protect against Omicron subvariants and to have improved efficacy against severe disease48. In addition, if intranasal vaccinations and nasal sprays become more developed, suppressing viral growth and clearing viruses in the nose and the upper respiratory system by triggering mucosal immune responses would deter viral spread49,50,51. To improve vaccine efficacy, the SARS-CoV-2 vaccine may need to be updated similar to the annual influenza vaccine.
References
Moore, S., Hill, E. M., Tildesley, M. J., Dyson, L. & Keeling, M. J. Vaccination and non-pharmaceutical interventions for COVID-19: a mathematical modelling study. Lancet Infect. Dis. 21, 793–802 (2021).
Ge, Y. et al. Untangling the changing impact of non-pharmaceutical interventions and vaccination on European COVID-19 trajectories. Nat. Commun. 13, 3106 (2022).
Oliu-Barton, M. et al. Elimination versus mitigation of SARS-CoV-2 in the presence of effective vaccines. Lancet Glob. Health 10, e142–e147 (2022).
Robinson, P. C. et al. COVID-19 therapeutics: challenges and directions for the future. Proc. Natl Acad. Sci. USA 119, e2119893119 (2022).
Shu, Y. & McCauley, J. GISAID: Global initiative on sharing all influenza data—from vision to reality. Euro Surveill. 22, 30494 (2017).
Telenti, A. et al. After the pandemic: perspectives on the future trajectory of COVID-19. Nature 596, 495–504 (2021).
Brosh-Nissimov, T. et al. BNT162b2 vaccine breakthrough: clinical characteristics of 152 fully vaccinated hospitalized COVID-19 patients in Israel. Clin. Microbiol. Infect. 27, 1652–1657 (2021).
Abu-Raddad, L. J. et al. Effect of mRNA vaccine boosters against SARS-CoV-2 omicron infection in Qatar. New Engl. J. Med. 386, 1804–1816 (2022).
Kuhlmann, C. et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet 399, 625–626 (2022).
Hirotsu, Y. et al. SARS-CoV-2 omicron sublineage BA.2 replaces BA.1.1: Genomic surveillance in Japan from September 2021 to March 2022. J. Infect. 85, 174–211 (2022).
Hirotsu, Y. & Omata, M. Detection of R.1 lineage severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with spike protein W152L/E484K/G769V mutations in Japan. PLOS Pathog. 17, e1009619 (2021).
Hirotsu, Y. & Omata, M. SARS-CoV-2 B.1.1.7 lineage rapidly spreads and replaces R.1 lineage in Japan: Serial and stationary observation in a community. Infect. Genet. Evol. 95, 105088 (2021).
Hirotsu, Y. & Omata, M. Discovery of a SARS-CoV-2 variant from the P.1 lineage harboring K417T/E484K/N501Y mutations in Kofu, Japan. J. Infect.82, 276–316 (2021).
Omata M, et al. The dynamic change of antibody index against Covid-19 is a powerful diagnostic tool for the early phase of the infection and salvage PCR assay errors. J. Microbiol. Immunolo. Infect. https://doi.org/10.1016/j.jmii.2020.12.009 (2021).
Ng, M.-Y. et al. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiol. Cardiothorac. Imaging 2, e200034 (2020).
Hani, C. et al. COVID-19 pneumonia: a review of typical CT findings and differential diagnosis. Diagn. Interv. Imaging 101, 263–268 (2020).
Shirato, K. et al. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn J. Infect. Dis. 73, 304–307 (2020). PubMed PMID: 32074516.
Hirotsu, Y. et al. Analysis of Covid-19 and non-Covid-19 viruses, including influenza viruses, to determine the influence of intensive preventive measures in Japan. J. Clin. Virol. 129, 104543 (2020).
Hirotsu, Y. et al. Direct comparison of Xpert Xpress, FilmArray Respiratory Panel, Lumipulse antigen test, and RT-qPCR in 165 nasopharyngeal swabs. BMC Infect. Dis. 22, 221 (2022).
Hirotsu, Y. et al. Prospective Study of 1,308 Nasopharyngeal Swabs from 1,033 Patients using the LUMIPULSE SARS-CoV-2 Antigen Test: Comparison with RT-qPCR. Int. J. Infect. Dis. 105, 7–14 (2021).
Hirotsu, Y. et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs including from 7 serially followed patients. Int. J. Infect. Dis. 99, 397–402 (2020).
Shepard, S. S. et al. Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genom. 17, 708 (2016).
Hadfield, J. et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 34, 4121–4123 (2018).
Rambaut, A. et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 5, 1403–1407 (2020).
Hansell, D. M. et al. Fleischner society: glossary of terms for thoracic imaging. Radiology 246, 697–722 (2008).
Salehi, S., Abedi, A., Balakrishnan, S. & Gholamrezanezhad, A. Coronavirus Disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. c 215, 87–93 (2020).
Ye, Z., Zhang, Y., Wang, Y., Huang, Z. & Song, B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur. Radiol. 30, 4381–4389 (2020).
Francone, M. et al. Chest CT score in COVID-19 patients: correlation with disease severity and short-term prognosis. Eur. Radiol. 30, 6808–6817 (2020).
Pan, F. et al. Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19). Radiology. 295, 715–721 (2020). PubMed PMID: 32053470.
Brogna, B. et al. COVID-19 pneumonia in vaccinated population: a six clinical and radiological case series. Medicina 57, 891 (2021).
Suzuki R, et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature https://doi.org/10.1038/s41586-022-04462-1 (2022).
Meng, B. et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 603, 706–714 (2022).
Nagakubo, Y. et al. Non-pharmaceutical interventions during the COVID-19 epidemic changed detection rates of other circulating respiratory pathogens in Japan. PLoS ONE 17, e0262874 (2022).
Nyberg, T. et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet 399, 1303–1312 (2022).
Tong, X. et al. Old age is an independent risk factor for pneumonia development in patients with SARS-CoV-2 Omicron variant infection and a history of inactivated vaccine injection. Infect. Drug Resist. 15, 5567–5573 (2022).
Fan, Y. et al. SARS-CoV-2 Omicron variant: recent progress and future perspectives. Sig. Transduct. Target.Ther. 7, 141 (2022).
Altarawneh, H. N. et al. Effects of previous infection and vaccination on symptomatic Omicron infections. New Engl. J. Med. 387, 21–34 (2022).
Andrews, N. et al. Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) Variant. New Engl. J. Med. https://doi.org/10.1056/NEJMoa2119451 (2022).
Peacock, T. P. et al. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. https://doi.org/10.1101/2021.12.31.474653 (2022).
Willett, B. J. et al. The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. https://doi.org/10.1101/2022.01.03.21268111 (2022).
Hui, K. P. Y. et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 603, 715–720 (2022).
Yamasoba D, et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell https://doi.org/10.1016/j.cell.2022.04.035 (2022).
Emanuel, E. J., Osterholm, M. & Gounder, C. R. A National Strategy for the “New Normal” of Life With COVID. JAMA 327, 211–212 (2022).
Suk, J. E. et al. Public health considerations for transitioning beyond the acute phase of the COVID-19 pandemic in the EU/EEA. Eurosurveillance 27, 2200155 (2022).
Levin, E. G. et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. New Engl. J. Med. 385, e84 (2021).
Yek, C. et al. Risk factors for severe COVID-19 outcomes among persons aged ≥18 years who completed a primary COVID-19 vaccination series — 465 Health Care Facilities, United States, December 2020–October 2021. MMWR Morb. Mortal Wkly Rep. 71, 19–25 (2022).
Offit, P. A. Covid-19 Boosters—Where from here? New Engl. J. Med. 386, 1661–1662 (2022).
Burki, T. COVID vaccine booster doses for Omicron variants. The Lancet Respir. Med. 10, 936 (2022).
Hassan, A. O. et al. An intranasal vaccine durably protects against SARS-CoV-2 variants in mice. Cell Rep. 36, 109452 (2021).
Lam, J.-Y. et al. A nasal Omicron vaccine booster elicits potent neutralizing antibody response against emerging SARS-CoV-2 variants. Emerg. Microb. Infect. 11, 964–967 (2022).
Balmforth, D. et al. Evaluating the efficacy and safety of a novel prophylactic nasal spray in the prevention of SARS-CoV-2 infection: a multi-centre, double blind, placebo-controlled, randomised trial. J. Clini. Virol. 155, 105248 (2022).
Acknowledgements
The authors would like to thank the clinical laboratory technicians in the microbiology laboratories at our institution. We also thank Katherine Thieltges from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript. This study was supported by a Grant-in-Aid for the Genome Research Project from Yamanashi Prefecture (to M.O. and Y.H.), the Japan Society for the Promotion of Science (JSPS) KAKENHI Early-Career Scientists JP18K16292 (to Y.H.), a Grant-in-Aid for Scientific Research (B) 20H03668 (to Y.H.), a Research Grant for Young Scholars (to Y.H.), the YASUDA Medical Foundation (to Y.H.), the Uehara Memorial Foundation (to Y.H.) Medical Research Grants from the Takeda Science Foundation (to Y.H.), and Kato Memorial Bioscience Foundation (to Y.H.).
Author information
Authors and Affiliations
Contributions
Y.H. drafted the manuscript, generated the genomic data, and performed the statistical analyses. Y.K. and A.S. collected and interpreted the clinical data. T.T., S.H., H.Y., S.I., M.K., and H.K. participated in the study and interpreted the clinical data. Y.M. collected data and supervised the study. M.O. conceptualized the study design and revised the manuscript. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Shu-Hsing Cheng, Ali Gholamrezanezhad and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hirotsu, Y., Kakizaki, Y., Saito, A. et al. Lung tropism in hospitalized patients following infection with SARS-CoV-2 variants from D614G to Omicron BA.2. Commun Med 3, 32 (2023). https://doi.org/10.1038/s43856-023-00261-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-023-00261-5