Abstract
Plant-derived phenolic acids are catabolized by soil microorganisms whose activity may enhance the decomposition of soil organic carbon (SOC). We characterized whether phenolic acid-degrading bacteria enhance SOC mineralization in forest soils when primed with 13C-labeled p-hydroxybenzoic acid (pHB). We further tested whether pHB-induced priming could explain differences in SOC content among mono-specific tree plantations in a 70-year-old common garden experiment. pHB addition primed significant losses of SOC (3–13 µmols C g−1 dry wt soil over 7 days) compared to glucose, which reduced mineralization (-3 to -8 µmols C g−1 dry wt soil over 7 days). The principal degraders of pHB were Paraburkholderia and Caballeronia in all plantations regardless of tree species or soil type, with one predominant phylotype (RP11ASV) enriched 23-fold following peak pHB respiration. We isolated and confirmed the phenolic degrading activity of a strain matching this phylotype (RP11T), which encoded numerous oxidative enzymes, including secretion signal-bearing laccase, Dyp-type peroxidase and aryl-alcohol oxidase. Increased relative abundance of RP11ASV corresponded with higher pHB respiration and expression of pHB monooxygenase (pobA), which was inversely proportional to SOC content among plantations. pobA expression proved a responsive measure of priming activity. We found that stimulating phenolic-acid degrading bacteria can prime decomposition and that this activity, corresponding with differences in tree species, is a potential mechanism in SOC cycling in forests. Overall, this study highlights the ecology and function of Paraburkholderia whose associations with plant roots and capacity to degrade phenolics suggest a role for specialized bacteria in the priming effect.
Similar content being viewed by others
Introduction
Forest soils are rich in plant-derived phenolic acids which represent a sizeable proportion of fast-cycling, low-molecular weight soil organic carbon (SOC),1 estimated at between 10 and 4000 mg · kg−1 dry organic layer soil.2,3,4,5 Concentrations of soil phenolics fluctuate as a function of plant inputs and phenolic acid-degrading activity.4,6,7,8,9,10 The cycling of soil phenolics has the potential to accelerate decomposition according to research linking phenolic acids, and phenol oxidase activity, to the soil priming effect.11,12,13,14,15,16 The priming effect refers to the enhanced decomposition of SOC which can occur when an influx of new C stimulates microbial activity, which is a fundamental process in terrestrial C-cycling.17,18 The activity of phenolic acid-degrading populations appear to be especially impactful, priming a greater loss of SOC (per unit biomass) than glucose or cellobiose.14 In this light, the low phenolic content of older accumulations of SOC19,20 may reflect a subsidence in the priming activity fueled by phenolics. Accordingly, we would expect to find a link between phenolic acid-degrading populations, soil priming, and SOC accrual in nature. However, this link has not yet been tested and the identity and ecology of these populations, and the mechanisms behind their priming activity, remain poorly described.
The primary sources of phenolic acids to soil are litter leachate, root exudates, and the decomposition products of lignin-rich plant residues. The composition of soil phenolics depends on the plant community, environmental conditions, and soil properties,21,22,23,24 with broad differences between coniferous and deciduous forests.2,25,26,27 p-hydroxybenzoic acid (pHB) is one of the most abundant phenolic acids found in plant litter, tree root exudates, and soil, ranging in concentrations from 0.1 to 50 mg · kg−1 dry wt in bulk soil4,5,7,28,29,30 which can be three to four-fold higher in the rhizosphere.31 Plant pHB content was found to correlate with the strength of priming13 and benzoic acid, an abundant root exudate with similar properties as pHB, can induce priming.15 To compare differences in pHB sources, and corresponding activity of pHB-degrading populations, we conducted an experiment using a 70-year-old common garden with plantations of a coniferous tree species (red pine) and two deciduous tree species: one leguminous (black locust) and one non-leguminous (sugar maple).32
Most phenolic acid-degrading microorganisms isolated from soil are fast-growing members of the Beta- and Gammaproteobacteria (Table S1).33,34,35,36 Rapid growth may be essential for metabolizing phenolics in soil, which quickly adsorb to surfaces of minerals and organic matter, reducing their bioavailability and promoting SOC accumulation.31,37,38,39 Though a culture-independent survey of phenolic acid-degrading microbes has not yet been made in forest soils, metagenomic analyses of lignin-degrading populations implicate Alphaproteobacteria (Rhizobiales, Sphingomonadales and Caulobacterales), Betaproteobacteria (Burkholderiaceae) and Actinobacteria (Nocardiaceae, Frankiaceae, Streptomycetaceae and Microbacteriaceae).40,41,42 Populations of Alpha- and Betaproteobacteria are abundant during white-rot decay of wood, where phenolic acid concentrations are high;43,44 and in the rhizosphere, where roots exude phenolic acids to facilitate plant-microbe interactions.45,46 Accordingly, we expect the phenolic-acid degrading populations that mediate priming will belong to these three bacterial classes, representing a narrower diversity of taxa than observed during glucose-induced priming.47
The priming effect can be generated by a variety of mechanisms that interact in a context-dependent manner.48,49 In certain cases, priming coincides with an increased oxidative potential of soils.11,12,50 Measures of potential oxidative activity may therefore serve as useful proxies for priming responses. pHB oxidation is catalyzed by p-hydroxybenzoate 3-monooxygenase (pobA; EC 1.14.13.2) as part of the peripheral pathways for the degradation of aromatics.51 PobA catalyzes an essential step in the degradation of phenolics, which can be the rate limiting step in the degradation of certain polyphenolics in soil.52 Furthermore, the expression of pobA is responsive to environmental conditions, evidenced by its upregulation in Burkholderia multivorans grown in soil versus laboratory media.53 Consequently, the expression of pobA may provide a proxy for the activity of phenolic acid-degrading soil populations and any potential priming effects.
In previous research, we have shown that an abundant aromatic acid in plant root exudates (benzoic acid) can prime decomposition mediated by the activity of phenolic-degrading bacteria.15,54 Our present study sought to test the link between phenolic acid-degrading populations, soil priming, and SOC accrual in a 70-year-old common garden experiment32 and the mechanism by which their metabolism controls soil priming. We tested whether pHB primes microbial activity that enhances SOC decomposition, and whether this phenomenon corresponded to differences in SOC content among monospecific tree plantations. We measured pHB degradation in the field and its capacity to prime SOC mineralization in a microcosm experiment. pHB-degrading populations were characterized using 16S rRNA amplicon sequencing, stable isotope probing (SIP) with 13C-pHB, the expression of pobA, as well as physiological and genomic characterization of pHB-degrading isolates. We hypothesized that the activity of pHB-degrading populations will prime the loss of SOC and that differences in this activity will correspond to variation in SOC content in forest soils. Specifically, we predicted (i) a positive relationship between the activity of pHB-degrading bacteria and SOC priming and (ii) an inverse relationship between SOC content in forest soils and pHB-degrading activity. Our microbe-centric approach supposes that the ecological and functional traits of phenolic acid-degrading bacteria are mechanistically important to soil priming and in environmental processes that govern the fate of SOC.
Material and methods
Description of field site
We studied pHB-degradation and the priming effect in soils from a 70-year old common garden experiment consisting of uniform 0.4 ha plots planted with monocultures of 13 tree species spanning types typical of northeastern USA (Turkey Hill Forest Plantation, Dryden, NY).32,55 The site was reforested following a period of over 100 years of agricultural use. We sampled plantations of sugar maple (“SM”, Acer saccharum Marsh; 42.451137, -76.420519), red pine (“RP”, Pinus resinosa Ait.; 42.450945, -76.420638), and black locust (“BL”, Robinia pseudoacacia L.; 42.451818, -76.420614) to capture variation in sources of soil phenolics. Plantations were chosen to include representatives of coniferous and deciduous tree species and legume-forming (black locust) and non-leguminous species (sugar maple). Trees were grown in typic Fragiochrept soils (inceptisols) belonging to either the Mardin B (“MaB”; Typic Fragiudepts) or Lordstown channery silt loam C soil series (“LnC”; Typic Dystrudepts). Our study examined a total of five combinations of tree species + soil type, which we have termed ‘ecoplots’, that encompassed a gradient in total soil organic matter: BLMaB (21.9% w/w) > SMMaB (14.5%) > BLLnC (14.0%) > RPLnC (12.75%) > SMLnC (10.2%). There were no plantations of RP in MaB soil. Earthworms populations, which can accelerate decomposition,56,57 did not correspond with trends in SOC in our plantations.58 Details on the site and three separate field sampling campaigns are provided in the Supplementary Methods. The concentration of bioavailable elements in soils was determined using the modified Morgan extraction procedure59 by the Cornell Nutrient Analysis Laboratory. Estimates of soil organic matter were made according to the loss-on-ignition method.
pHB-induced soil priming
Microcosms were prepared with soils from BLLnC, RPLnC and SMLnC to test whether pHB degrading activity induced priming and to identity and link active populations with those observed in the field experiment. Microcosms were prepared and amended with the following treatments: (i) water-only control, (ii) 13C-glucose, (iii) 13C-pHB, (iv) 13C-pHB + PobA inhibitor (4-hydroxy-3-iodobenzoate) and (v) a PobA inhibitor-only control (overview in Fig. S1). The inhibition of PobA was performed to measure the direct contribution of PobA activity to priming. An additional set of microcosms was used to test whether priming could be induced by artificially increasing the abundance of a phenolic acid-degrading isolate (RP11T), corresponding to main pHB-degrading phylotype in field data (RP11ASV; details in Supplementary Methods). Microcosms were prepared using fresh soil collected from the upper 5 cm of a 20 × 20 cm sampling plot, which was sieved to remove root and litter (2-mm), homogenized, and pre-incubated for 10 days. Sieving, homogenization, and pre-incubation were necessary to reduce soil SOC heterogeneity and limit variability in the concentration of labile SOC at the time of sampling. After pre-incubation, soils were air dried for 48 h in a sterile biosafety hood. Ten grams of dry soil were added to 120-mL serum vials and wetted during treatment application to 60% water holding capacity. Glucose and pHB were added at a concentration of 0.5 mg C per g dry wt soil. Each treatment was run in quadruplicate per ecoplot. Isotopically labeled substrates (17.5 atom % 13C) or RP11T cells (99 atom % 13C) were used to distinguish between the CO2 derived from amendment versus SOC. The isotopic composition of amendments was confirmed by EA-IRMS by the Cornell Stable Isotope Laboratory. Headspace sampling was performed every 24 hrs for a week with the headspace exchanged with filtered air after each sampling. The quantity of 12CO2 (m/z 44) and 13CO2 (m/z 45) was analyzed using GC/MS (Shimadzu GCMS-QP2010S) with a set of standards ranging from 1000 to 40,000 ppm CO2. After 7 days, sub-samples of 0.5 g of wet wt soil were stored at −80 °C for RNA-based quantification of pobA gene abundance and bacterial community composition. Headspace CO2 measurements continued at reduced frequency for a total of 26 days. All methodological details are available in the Supplementary Methods.
pHB degradation in field soil
To test whether phenolic acid-degrading activity differed among ecoplots, pHB mineralization rates were measured in field soils in an experiment which also served to identify the active in situ populations via DNA-SIP. Sampling took place in three randomly selected plots (20 × 20 cm) per ecoplot. The litter layer was cleared to expose the Oa / A-horizon soil and stainless-steel chambers (radius 1.25 cm) were driven into the soil at an even depth (2 cm). Photographs of field sampling are provided (Fig. S2). A thin layer of surface soil was evenly wetted by dripping 150 µl of either 13C7-labeled p-hydroxybenzoic acid (99% atom 13C; Sigma-Aldrich) or unlabeled control (‘12C’; Sigma-Aldrich) at a concentration of 1000 ppm (6.9 and 7.3 mM, respectively). Chambers were immediately enclosed with rubber septa and 13CO2 concentrations were measured in the headspace at five sampling timepoints over a period of 27 h. At each time point, 2.5 mL of headspace was sampled and stored in evacuated 2-mL vials prior to analysis with a Hewlett Packard 5890 gas chromatograph (Wilmington, DE) equipped with a 5971A mass selective detector (details in Supplementary Methods). Net 13CO2 was determined by subtracting average natural abundance 13CO2 in 12C controls from the total 13CO2 respired in 13C-pHB-amended soils. Following the incubation period, ~0.5 g of soil was collected from the upper 1 cm of the dosed soil, transported on ice and stored at −80 °C for use in DNA-SIP.
Identifying pHB-degrading populations
Nucleic acids were extracted from 0.5 g of soil using the modified phenol-chloroform method from60 following bead-beating at 5.5 m·s−1 for 2.5 min (Bio Spec Products, Santa Clara, CA, USA) in 2-mL Lysing Matrix E Tubes (MP Biomedicals, OH). For the DNA-SIP experiment, DNA was extracted from 13C- and 12C-pHB amended field soils and 13C-enriched DNA was separated from unlabeled DNA by CsCl density gradient ultracentrifugation as previously described.61,62 Twenty 200-µL fractions were collected from each CsCl gradient and 16S rRNA gene amplicon sequencing libraries were prepared for fractions F1-2 (pooled), F3, F4, F5, F6, F7, F8, F9-10, and F11-12 (spanning CsCl buoyant density of 1.77–1.70 g·mL-1). RNA was coextracted with DNA for quantifying the expression of pobA, in field soils and microcosms, and for profiling the active populations in microcosms targeting 16S rRNA. Complementary DNA (cDNA) was synthesized from DNAse-treated RNA extracts using SunScript reverse transcriptase (Expedeon, San Diego USA). Bacterial community composition was assessed by amplification of the V4 region using polymerase chain reaction (PCR) with dual-indexed barcoded 515f/806r primers.63 PCR was performed in duplicate and pooled, and PCR products were purified and normalized to a standard concentration prior to sequencing on three lanes of Illumina MiSeq (2 × 250 paired end) at the Cornell Biotechnology Resource Center. Amplicon libraries were deposited in the European Sequence Archive under the BioProject accession PRJEB23740. Following sequencing processing (described below), amplicon sequence variants (“phylotypes”) were designated as pHB-degraders if their relative abundance was at least eightfold greater in heavy CsCl fractions (F3-F8) in 13C-enriched versus 12C-control amplicon libraries, after variance stabilization by DESeq2 (v. 1.24.0).64 Phylotypes with increased relative abundance in rRNA pools in the microcosm-based priming experiment were identified by indicator species analyses using the R package “indicspecies”.65 Full details on methods and controls for DNA extractions, cDNA synthesis, PCR conditions and isopycnic gradient centrifugation are available in the Supplementary Methods.
Isolation, genome sequencing, and functional description of pHB degraders
pHB degrading isolates were obtained from red pine and sugar maple LnC soil by serial dilution. Diluted cells were plated onto mineral salts media containing 3 mM pHB (MSM-pHB) as the sole carbon source and incubated at room temperature. Colonies appeared after 3 days and were isolated by streaking on MSM-pHB and stored in 20% glycerol (v/v) at −80 °C. Genomic DNA was extracted from isolates and isolates were identified by Sanger sequencing of the 16S rRNA gene (27f/1492r).66 The pHB degrading activity of isolates was validated based on protocatechuate production with GC/MS (details in Supplementary Methods). An isolate, RP11, matched one of the predominant pHB-degrading phylotypes in amplicon libraries and its genome was sequenced using a quarter lane of Illumina MiSeq (2 × 250bp). The sequencing reads and genome assembly are available from the NCBI (BioProject PRJNA558488). The strain was subsequently described as Paraburkholderia madseniana RP11T (DSM 110123) and exhibited growth on several phenolic acids including p-coumaric and phthalic acids.54 Details on the assembly, calculation of average nucleotide identity (ANI) and functional gene annotation are available in the Supplementary Methods.
Quantifying pobA expression
Custom PCR primers were designed to target a 75-bp fragment of the “pobA1” gene identified in the genome of isolate RP11T: pobA1_f (5’-AGA TCG AAT CCA CCA TCC GC-3’) and pobA1_r (5’-TTC AAA GCC GTG ATG CAA CG -3’) using NCBI’s Primer-BLAST software.67 Details of primer design and validation and RT-qPCR conditions are provided in the Supplementary Methods. In brief, the quantification of pobA and 16S rRNA transcripts (primers 341f/534r) was performed by RT-qPCR using an AB7300 Real-Time PCR system (Applied Biosystems).68 pobA gene expression was assayed using triplicate soil cores per ecoplot in the field. Cores were taken with 3-mL plastic syringe barrels and immediately extruded into 10-mL serum bottles and dosed with 75 µL of 13C-labeled pHB (1000 ppm). Expression levels were determined in RNA extracted from 1 g of soil following a 24-h incubation in field and following a 7-day period in microcosms.
Prevalence of pobA in environmental metagenomes and bacterial genomes
To gain insight into the diversity of bacteria capable of pHB-degradation and to identify environments with the greatest potential activity, we conducted surveys of the prevalence of pobA in publicly available metagenomic (n = 14,139) and genomic datasets (n = 11,643) through the IMG/ER portal.69 The number of pobA copies per genome and the relative abundance in metagenomes were determined using the KEGG function search tool (March 19th, 2019). Taxonomic classifications were based on those provided by IMG/ER and the environmental source of metagenomes was attributed to the ‘habitat’ descriptor. Custom pobA Hidden Markov Models (HMMs) were generated from an alignment of homologs present in all Burkholderiaceae genomes in the NCBI refseq_genomic database (n = 3991; as of June 20th, 2019). HMMs were developed for each of the seven pobA clades. A detailed description of all pobA analyses are provided in the Supplementary Methods and Supplementary Results, including information on enzyme structure and function and the phylogenetic trait depth of pobA paralogs (Table S2 and S3).70
Data analysis and statistics
Amplicon libraries were quality processed using QIIME2 (v. 2017.9)71 with dependency on DADA2 (v. 1.2.0)72 to assign sequences to amplicon sequence variants (ASVs), referred to throughout as phylotypes. Taxonomic classification was performed using the QIIME2 “q2-feature-classifier” trained on the Silva database (nr_v132).73 Libraries were filtered to remove phylotypes present in reagent blanks or in low relative abundance (< 0.05% of all sequences) or few samples (minimum three samples). Estimates of species richness and Pielou’s evenness were calculated using rarified data (nmin = 9140 per sample). All other analyses were performed on counts normalized to library totals (counts per thousand). All statistical analyses were performed in R (v. 3.4.0; R Core Team, 2018) with general dependency on the following packages: ggplot2 (v. 3.2.1)74 and phyloseq (v. 1.22).75 Variable selection was performed to identify the soil physicochemical parameters contributing most to betadiversity was performed using stepwise regression with the “step” function in R. Non-metric multidimensional scaling (NMDS) and perMANOVA were performed with parameters that contributed the most to variation using the R package “vegan” (v. 2.5-6).76 Betadiversity was calculated for whole amplicon libraries using Unifrac distances based on a rooted tree produced using the QIIME2 wrapper “q2-phylogeny” for MAFFT (v. 7.407)77 and FastTree (v. 2.1.10).78 Selected parameters were then fit to the NMDS with the ‘envfit’ function in vegan. The occurrence of Paraburkholderia in studies of phenolic acid-induced priming, root associations and lignin decomposition was determined by re-analyzing published 16S rRNA gene data using the aforementioned workflow (see Supplementary Results). All analyses are reproducible using R scripts in the Supplementary Data.
Results
We measured the priming effect in soil microcosms using 13C-labeled substrates (pHB and glucose) to differentiate the respiration of substrate from native SOC, while monitoring changes in microbial community composition and pobA expression. The activity and composition of pHB-degrading bacteria were determined in field soils by monitoring pHB respiration, pobA expression and performing DNA-SIP. This combined approach enabled us to link pHB-degrading populations with priming activity and track their activity in the 70-year-old common garden plantations (“ecoplots”) which differed by tree species, soil type, and SOC content. Differences in the sources and composition of phenolics, according to tree species, were expected to promote variation in phenolic acid-degrading/priming activity and community structure, which was expected to reflect differences in total SOC content.
pHB-induced soil priming in microcosms
The addition of pHB primed the decomposition of significant amounts of SOC during the first four days of incubation, when rates of pHB respiration were highest (Fig. 1A). The majority of pHB had been mineralized by day 7 (Fig. 1), corresponding with the cumulative priming of 3.3 (SMLnC), 5.5 (BLLnc), and 13 (RPLnC) µmols C g−1 dry wt soil, representing 3.6%, 4.5%, and 8.7% more SOC mineralization than soils that received water only. In contrast, glucose reduced SOC mineralization between −3 to −8 µmols C g−1 dry wt soil in the same period. pHB degradation and priming were significantly higher in RPLnC than BLLnC and SMLnC (TukeyHSD; p < 0.05), while differences between BLLnC and SMLnC were not significant. pHB was respired more rapidly and to a greater extent (93–100% of amendment) than glucose (62–71%) in all ecoplots (Fig. 1B). Inhibition of pobA, while effective in pure culture (see Supplementary Results), was ineffective in soil, having no effect on pHB respiration or pHB-induced priming in soil microcosms (Fig. 1).
The addition of pHB caused positive priming (A), which corresponded with maximal respiration of 13C-pHB (B) during the first 7 days of incubation (shaded in gray). Primed SOC is a measure of the native SOC respired relative to water-only controls. Error bars correspond to standard deviations among replicates (n = 4). An asterisks denote time points where respiration of native SOC significantly differed from water-only control. Priming was assessed in LnC soils which varied in soil organic matter content (BLLnC: 14.0% > RPLnC: 12.7% C > SMLnC: 10.2%).
In soil microcosms, 70 bacterial phylotypes increased in relative abundance in response to glucose (n = 8), pHB (n = 10), water & glucose (n = 36) or glucose & pHB (n = 16; Table S4). Several phylotypes classified to Paraburkholderia and Rhodanobacter were consistently enriched in all microcosms supplied with pHB and glucose, respectively (Fig. 2; Table S4). Four of the seven phylotypes with the greatest response to pHB (relative abundance in pHB versus water-only) were Paraburkholderia, while the others were Alicyclobacillus (SMLnC), Streptacidiphilus (RPLnC) and an unclassified member of the phylum WPS-2 (RPLnC). The relative abundance of Paraburkholderia increased in response to pHB and glucose in all soils (Fig. 2), though this effect was not significant in BLLnC. In addition, Paraburkholderia responded more strongly to pHB than to glucose in SMLnC soil (TukeyHDS; p < 0.01; Fig. 2). Transcripts of pobA were undetectable at day 7 (detection limit ~300 copies per µL; data not shown) consistent with pHB mineralization dynamics (Fig. 1A).
Soils amended with glucose, or with RP11T cells, mineralized less SOC than those that received only water (i.e., “negative priming”), though the effect was statistically significant only in the latter treatment (Fig. S3). Amendment with RP11T cells produced 5–14% less SOC mineralization when compared to controls. In all cases, negative priming was associated with steep increases in the relative abundance of Streptomyces and Streptacidiphilus phylotypes (Fig. S3; details in Supplementary Results).
pHB-degrading activity of field soils
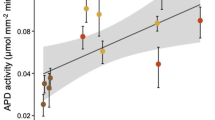
The respiration of pHB and expression of pobA were compared among ecoplots to test whether tree species (phenolic source) and soil type affected pHB-degrading activity. The amounts of pHB respired in situ differed considerably among ecoplots (Fig. 3). pHB was respired rapidly in SMLnC and RPLnC soils with maximum instantaneous rates observed after about 10 h (Fig. 3A, B). In contrast, pHB-degrading activity was relatively modest in SMMaB, BLMaB and BLLnC, with substrate respiration plateauing at ~1.5–2% of the total C added to soil (Fig. 3B). SOC content was lowest in SMLnC and RPLnC where the highest rates of pHB respiration, pobA transcription, and the highest relative abundances of RP11ASV occurred (Fig. 3C, D, E). These trends were not driven by total microbial biomass since RNA yield and 16S rRNA abundance were directly proportional to soil organic matter content (Fig. S4A, B).
In situ measurements show that maximal rates of pHB respiration (A, B) are associated with high relative abundance of the RP11 phylotype (C), high levels of pobA transcripts (D), and low levels of soil organic matter (E). Error bars correspond to standard deviation (n = 3) and letters denote significant differences (p < 0.05) based on Tukey’s Honest Significant Difference tests (n = 3).
pHB-degrading populations
The active pHB-degrading populations were identified by 16S rRNA gene sequencing in field soil by DNA-SIP and in microcosm with RNA sequencing. Overall bacterial community composition in the field varied most by tree species (R2perMANOVA = 0.25; p < 0.001) followed by soil type (R2 = 0.14; p < 0.001), illustrated by the NMDS ordination (Fig. 4A). Of all the soil properties, pH and aluminum content explained the most variation in community structure (R2perMANOVA = 0.096 and 0.07, respectively; Table S5). BL forest soils had significantly lower pH, higher iron and sulfur, and lower cadmium and manganese than other forest types, while LnC and MaB soil types differed by aluminum and calcium content (Fig. 4B; Table S6). These data show that major differences in soil properties and microbial communities had developed among ecoplots during the 70-year period since planting.
NMDS ordination based on Bray-Curtis dissimilarity of 16S rRNA sequences from pHB-amended field soils indicate that soil series, tree species, and soil properties influence community composition (A). Soil physicochemical properties differed among ecoplots (B). In (A), community composition is represented by the average of all gradient fractions for 13C-labeled and 12C-control libraries for each ecoplot (n = 6). In (B), letters denote significant differences in soil properties (p < 0.05) based on Tukey’s Honest Significant Difference tests (n = 3). The hatching of box plots denotes the Mardin soil type. Measured concentrations correspond to the bioavailable elemental content of soils according to the modified Morgan extraction procedure. Comprehensive soil physicochemical properties are available in Table S6.
Despite these differences, we observed that the same six Burkholderiaceae phylotypes dominated in situ pHB degradation in all ecoplots (Fig. 5A). Bacterial communities profiled from the 13C-labeled DNA from heavy CsCl gradient fractions exhibited greatly reduced richness and evenness relative to unlabeled DNA, indicating strong 13C-enrichment of pHB-degrading subpopulations (Fig. S5). Between 10 and 40% of all amplicon sequences in 13C-DNA pools corresponded to phylotypes classified as Paraburkholderia (n = 8) and Caballeronia (n = 4), of which six predominated in all ecoplots (Fig. 5A, B). Most of these phylotypes were also enriched by pHB addition in the microcosm priming experiment, including four of the six major phylotypes (Table S7). Two Paraburkholderia phylotypes, matching P. madseniana (RP11ASV) and P. xenovorans LB400, were the dominant pHB-responders in both soil microcosm priming experiments and DNA-SIP experiments performed in field soils (Figs. 2 and 5).
Paraburkholderia represented one-third of all phylotypes enriched significantly in 13C-DNA as determined by their log2-fold differential abundance relative to corresponding 12C-DNA controls shown in (A). Paraburkholderia and Caballeronia phylotypes identified in (A) comprised the seven most relatively abundant phylotypes in 13C-DNA, and these phylotypes were found in all tree plantations, as shown in (B). The two most abundant 13C-labeled phylotypes matched P. madseniana RP11T and P. xenovorans, respectively (B). Bars are colored by taxonomic classification and labeled with an abbreviated taxon name where bar coloring is obscured. Error bars correspond to standard deviation (n = 3). Phylotypes were designated to a species group based on the top representative BLAST hit to a type strain (100% identity). Details on the rank and relative abundance of phylotypes are available in Table S7.
Less abundant 13C-pHB-labeled phylotypes detected in the DNA-SIP experiment included Burkholderia, Collimonas, Pseudomonas (Gammaproteobacteria), Phenylobacterium (Alphaproteobacteria), Streptomyces and Conexibacteraceae (Actinobacteria), and Solicoccozyma (a basidiomycotal yeast; Fig. S6). These phylotypes were observed sporadically among forest and soil types (Fig. 5B; Table S7), unlike phylotypes of Paraburkholderia and Caballeronia which predominated following pHB addition in all ecoplots.
Isolation of pHB-degrading bacteria
Nine bacteria were isolated from RPLnC soil using minimal media with pHB as the sole carbon source. All of these isolates produced protocatechuate (the reaction product of PobA) during growth on pHB (data not shown). Isolates were classified as Paraburkholderia (n = 5), Pseudomonas (n = 3) and Cupriavidus. Four of the Paraburkholderia isolates had identical full-length 16S rRNA genes that matched one of the dominant pHB-degrading phylotypes (RP11ASV) detected in DNA-SIP and microcosm experiments. One of these strains, isolate RP11T, was chosen for genome sequencing. The RP11T genome was large (10.1 Mb), contained two chromosomes and encoded 6 copies of the rrn operon.54 The genome encoded the complete pathway to mineralize pHB and included two paralogs of pobA which shared 48% amino acid sequence identity. RP11T encoded an array of oxidative enzymes, including a DyP-type peroxidase, laccases, aryl-alcohol oxidases and an array of ring-cleaving and ring-hydroxylating dioxygenases (Table S8). Several of these enzymes contained signal peptide sequences, indicating potential for extracellular activity, including the DyP-type peroxidase, three laccases, and an aryl-alcohol oxidase.79
Phylogenetic diversity and environmental distribution of pobA-encoding bacteria
In the IMG-ER genome database, homologs of pobA were most commonly encoded in genomes of Betaproteobacteria and Alphaproteobacteria, occurring in more than 45% of genomes classified to these groups (Fig. 6A, and Table S9). Genomes encoding paralogs (two copies of pobA) were more common in Paraburkholderia (25% of genomes) than any other genus of pobA-encoding Burkholderiaceae (Fisher’s exact, O.R. = 3.1, p < 0.001). In addition, pobA paralogs were more common in soil isolates compared to other environmental sources (O.R. = 2.7, p < 0.001; Fig. S7A). Furthermore, pobA homologs were more abundant in forest soil metagenomes than in other environmental sources, and this difference was significant (Fig. 6B). Homologs of pobA comprised seven major phylogenetic clades, and the two largest of these clades were represented by pobA1 and pobA2 of RP11T (clades 6 and 5, respectively, Fig. S7B). Additional information on the phylogenetic diversity and inferred activity of pobA homologs can be found in the Supplementary Results.
The phylogenomic distribution and environmental associations of pobA in (A) bacterial genomes (n = 11,643) and (B) environmental metagenomes (n = 14,139) from the IMG/ER database. In (A), phyla were ranked by the proportion of genomes encoding pobA. Only phyla represented by >10 genomes were surveyed, and only those with at least one genome encoding pobA are shown. The taxonomic classification of pobA-encoding genomes from Alpha- and Betaproteobacteria are indicated by the inset pie charts in (A). Each taxonomic group was normalized to the total number of genomes present in the database. Homologs of pobA were far more common in soils in general, and in forest soils in particular, relative to other sample types as shown in (B). Letters denote significant differences (p < 0.05) based on pairwise Wilcoxon Rank Sum tests.
Discussion
Our results demonstrate that the degradation of p-hydroxybenzoic acid (pHB) can prime the decomposition of SOC in a variety of forest and soil types. This phenomenon was driven by the activity of Paraburkholderia and, to a lesser extent, Caballeronia, from the family Burkholderiaceae. The predominant Paraburkholderia phylotypes were related to P. madseniana RP11T and P. xenovorans LB400T, species noted for having more genes and pathways for the degradation of phenolics and aromatics than close relatives,54 and among the highest of any bacterial species.80 Based on these observations, we conclude that the mechanisms underlying phenolic- and glucose-induced priming differ, since the former is driven by a closely related group of bacteria with marked functional specialization and the latter by the general activity of diverse populations.47 Repeated dosing of glucose is commonly required to cause priming,47 with a single bolus often resulting in negligible or negative priming, consistent with our observations.15,81,82,83 In contrast, a single dose of pHB (Fig. 1), benzoic acid15 or vanillin14,16 resulted in positive priming and the enrichment of Paraburkholderia in forest, agricultural and tundra soils, respectively. The identification of the taxa and functional genes responsible for phenolic-induced priming is an important step forward in understanding the types of microbial activity responsible for enhancing losses of SOC. Indeed, several lines of evidence in our study suggest phenolic acid-degrading bacteria play a general role SOC cycling, such as the correspondence between SOC content among tree types in our common garden and rates of phenolic acid-degrading activity.
Characteristics of pHB-induced soil priming
The catabolism of pHB and glucose differed in ways that revealed differences in the nature of priming. The addition of pHB to soil yielded significant positive priming with nearly twice the amount of substrate mineralized compared to glucose, which yielded slight, though insignificant, negative priming (Fig. 1). This difference in substrate use efficiency has been documented in forest soils, where added pHB was mineralized at twice the rate of glucose, and produced only half the microbial biomass.84 The same effect occurred in grassland soil amended with vanillic acid, which yielded higher mineralization rates and lower microbial biomass than glucose or cellobiose, and also a greater positive priming effect by energy content.14 We hypothesize that pHB-induced priming is caused, in part, by a stoichiometric imbalance whereby C catabolism greatly exceeds the needs of anabolism, evident in the high rates of pHB mineralization. Stoichiometric limitation-induced degradation of soil organic matter is a well-known mechanism in priming.47,49,81,85,86 This could also explain why the turnover of RP11T cells (a relatively rich nutrient source) was associated with strong suppression of SOC mineralization (Fig. S3). We propose that variation in the priming effect can be governed by substrate-specific metabolic use efficiency, due to differences in stoichiometric imbalances promoted during metabolism. These dynamics are governed by life-history traits of decomposer populations or competitive and facilitative interactions with other soil microbes which are not yet fully understood.
Growth rate is a common determinant of substrate use efficiency and, in our case, may contribute to the high mineralization rates of pHB. pHB-degrading populations were dominated by fast-growing bacteria which tend to be less efficient in converting substrate C into biomass.87 Members of the family Burkholderiaceae are major zymogenous populations in soil35 and the rapid in situ growth of RP11ASV was evident in its increase from ~0.7 to 15% of total sequences in 24 h, during the period of maximal pHB respiration. Consistent with these observations, isolate RP11T exhibited rapid growth (µ = 0.22 h−1) in pure culture with pHB as the sole carbon source,54 comparable to the specific growth rate of E. coli on acetate (µ = 0.18 h−1).88 The RP11T genome also encoded six copies of the rrn operon, a characteristic of high growth rate.89 These observations suggest that rapid growth is a microbial trait relevant to priming, highlighting the importance of pulsed sources of phenolic C in SOC cycling.
Priming was also contingent on the metabolic state of degrader populations. Identical Paraburkholderia phylotypes responded to both glucose and pHB additions in RPLnC and BLLnC (Fig. 2), yet positive priming was induced only by pHB. This phenomenon was apparent in a related study on soil priming, where the same Paraburkholderia phylotypes responded to benzoic acid and glucose amendment, yet only benzoic acid elicited positive priming.15 Notably, the co-addition of glucose and benzoic acid enhanced priming and coincided with an even greater increase in Paraburkholderia (identical to our two major phylotypes). These observations are supported by the regulation of oxidative catabolic pathways in Paraburkholderia by aromatic and phenolic compounds, which are insensitive to glucose concentration.90,91 We conclude that the regulation of metabolic state is critical for priming by phenolic-degrading populations, which may be enhanced, but not triggered, by glucose or other non-inducing substrates (Fig. 7).
In (A), limiting SOM priming occurs when phenolic acid-degrading bacteria are rare and aromatic metabolism is not induced. In (B), the addition of non-inducing substrates, such as glucose, stimulate growth of aromatic acid-degrading bacteria, but priming does not occur if the concentration of aromatic acids is insufficient to induce EOE. In (C), the addition of inducing substrates, such as phenolic or other aromatic acids, stimulate SOM priming by inducing EOE and by promoting the growth of specialized degrader populations. In (D), if the concentration of endogenous aromatic acids is sufficient to induce EOE then the co-addition of non-inducing substrates can enhance SOM priming by promoting the growth of phenolic acid-degrading bacteria. Hence, while glucose can stimulate the growth of Paraburkholderia, priming only occurs when aromatic metabolism is induced. In (E), plant roots gain access to inaccessible forms of N and P by stimulating the growth and induce priming activity of phenolic acid-degrading bacteria by exuding phenolic/aromatic acids.
The activity of oxidative enzymes has long been considered a potential mechanism in priming, resulting from the co-metabolism of SOC.11,81 Paraburkholderia are renowned for their oxidative degradation of aromatics, phenolics, and polyaromatic hydrocarbons.92,93 The two dominant pHB-degrading phylotypes in our study matched to P. madseniana and P. xenovorans, species which encode an extensive array of aromatic degrading pathways.54,94 P. madseniana RP11T encoded among the greatest number of aromatic degrading genes compared to close relatives, including several putatively secreted oxidases79 and aryl-alcohol oxidases important for bacterial lignin degradation.42 P. madseniana RP11T also grows on phthalic acid, a major by-product of lignin degradation.95,96,97 Indeed, our retrospective analysis found that the two major pHB-degrading phylotypes of Paraburkholderia predominated in studies of lignin degradation42, and white rot decay43, and were the principal phylotypes in all phenolic/aromatic-based studies of soil priming14,15,16 (see Supplementary Results). All evidence indicates that Paraburkholderia, and their close relatives, cause priming by monopolizing phenolic acid degradation and co-metabolizing SOC.
Impacts of phenolic acid-degrading populations on soil C-cycling
Paraburkholderia and Caballeronia routinely occur where lignin- and phenolic-rich plant matter is decomposed,43,44,98,99,100 and in association with plant roots,101 where aromatic- and phenolic-rich exudates can recruit Paraburkholderia during development.102,103 The capacity for phenolic acids to prime decomposition, and abundance of plant-derived phenolic sources to soil, raises questions about the potential impact of this phenomenon on soil C-cycling. Phenol oxidase activity and SOC content are inversely correlated at ecosystem scales, but this relationship is weak and highly interrelated with other soil properties.104 In our field experiment, pHB-degrading activity and total SOC content were inversely proportional. pHB-degrading activity was greatest where the relative abundance of Paraburkholderia and pobA expression were highest, and where SOC was lowest (SMLnC and RPLnC). These observations raise the prospect that phenolic-induced priming may influence forest SOC cycling at scale. However, we interpret these results with caution, given that a single time point cannot capture the full extent of processes contributing to differences in SOC in our 70-year-old common garden. SOC content also followed trends in soil pH, iron, nickel and arsenic content which could independently affect SOC accrual. Still, the potential for plant-derived phenolics to influence SOC formation via priming is notable, given the biology and ecology of the phenolic acid-degrading populations.
Paraburkholderia and Caballeronia can exist as endophytes in a broad range of plants105,106,107,108,109,110 and are capable of nodulating legumes, including black locust.111 All three of the closest related genomes to RP11T were isolated from plant roots: P. sycomorum ST111 (94.3% ANI; from Acer roots)112, P. aspalathi LMG 27731T (94.2%; Aspalathus root nodule)113 and P. sp. OK233 (94%; Populus roots).114 The capacity to form associations with roots suggests that rhizosphere stimulation may underlie differences in pHB-degrading activity among tree types and why differences in pHB-degrading activity were observed in situ and not in microcosms (i.e., absent of root influence). The highest in situ pHB-degrading activity was recorded in SM plantations where root density was by far greatest, according to field observations (Fig. S1) and prior research.32 In contrast, soils in BL plantations exhibited the lowest pHB-degrading activity and were comprised of different Burkholderiaceae populations (Fig. 2). Black locust was the only leguminous tree species in our common garden capable of obtaining nitrogen through internal root symbioses, indicating differences in phenolic acid-degrading activity and priming may result from different relationships between plant and microbe. Phenolic root exudates are known to facilitate plant-microbe interactions that can be essential for plant nutrition.45,115 For instance, pHB concentrations doubled in exudates of poplar trees grown in nitrogen and phosphorus limiting conditions.31 We hypothesize that certain tree species may modulate the phenolic acid content of exudates to stimulate priming and gain access to nutrients stored in soil organic matter (Fig. 7E).
Tracking priming activity with pobA
The expression of pobA was consistent with pHB-degrading activity and reflected the transient nature of pHB-induced priming. Tracking priming activity based on the expression of functional genes can discern between mere demographic changes in populations associated with priming and the metabolic states associated with real priming activity. In our study, the relative abundance of Paraburkholderia increased when either glucose or pHB were added but priming occurred exclusively in the latter instance. Changes in the relative abundance of pobA in soil metagenomes was also coincident with vanillin-induced priming, and an increase in the P. xenovorans phylotype.16 Together, these findings illustrate the potential utility for tracking priming dynamics using pobA—a key enzyme in the peripheral degradation of phenolics.51
Conclusions
We conclude that specialized bacteria from the family Burkholderiaceae act as principal agents of phenolic acid-induced soil priming in forest soils, and likely more broadly. These populations consisted primarily of fast-growing Paraburkholderia characterized by their capacity to produce strongly oxidative catabolic enzymes, and to associate with plant roots. Our study builds on our prior work showing the priming activity of Paraburkholderia,15 and characterizing the predominant phenolic acid degrading isolate as a new species,54 by providing the essential microbiological evidence for understanding their ecology and function in priming. The transient nature of pHB-induced priming underscores the importance of pulsed resource availability, or “hot moments”, in soil C-cycling,116 and the utility of dialing in on functional gene expression. Moving forward, it is imperative to characterize the role of phenolic acid-degrading activity and priming in natural settings where seasonal and root-mediated dynamics are considered. Even slight environmental changes can lead to changes in priming, as evidenced by the effect of warming on phenolic acid-induced degradation of recalcitrant SOC by Paraburkholderia in tundra soil.16 Future studies of phenolic acid-induced priming might help shed light on the nature of SOC persistence, which has previously been linked to low phenolic acid content.19,20
References
Van Hees, P. A. W., Jones, D. L., Finlay, R., Godbold, D. L. & Lundström, U. S. The carbon we do not see - The impact of low molecular weight compounds on carbon dynamics and respiration in forest soils: a review. Soil Biol. Biochem. 37, 1–13 (2005).
Shindo, H., Ohta, S. & Kuwatsuka, S. Behavior of phenolic substances in the decaying process of plants: IX. Distribution of phenolic acids in soils of paddy fields and forests. Soil Sci. Plant Nutr. 24, 233–243 (1978).
Katase, T. Distribution of different forms of p-hydroxybenzoic, vanillic, p-coumaric and ferulic acids in forest soil. Soil Sci. Plant Nutr. 27, 365–371 (1981).
Muscolo, A. & Sidari, M. Seasonal fluctuations in soil phenolics of a coniferous forest: effects on seed germination of different coniferous species. Plant Soil. 284, 305–318 (2006).
Whitehead, D. C., Dibb, H. & Hartley, R. D. Bound phenolic compounds in water extracts of soils, plant roots and leaf litter. Soil Biol. Biochem. 15, 133–136 (1983).
Kuiters, A. T. & Sarink, H. M. Leaching of phenolic compounds from leaf and needle litter of several deciduous and coniferous trees. Soil Biol. Biochem. 18, 475–480 (1986).
Gallet, C. & Pellissier, F. Phenolic compounds in natural solutions of a coniferous forest. J. Chem. Ecol. 23, 2401–2412 (1997).
Schofield, J. A., Hagerman, A. E. & Harold, A. Loss of tannins and other phenolics from willow leaf litter. J. Chem. Ecol. 24, 1409–1421 (1998).
Kaiser, K., Guggenberger, G., Haumaier, L. & Zech, W. Seasonal variations in the chemical composition of dissolved organic matter in organic forest floor layer leachates of old-growth Scots pine (Pinus sylvestris L.) and European beech (Fagus sylvatica L.) stands in northeastern Bavaria, German. Biogeochemistry. 55, 103–143 (2001).
Li H. et al. Forest gaps alter the total phenol dynamics in decomposing litter in an alpine fir forest. PLoS ONE. 11, e0148426 (2016).
Blagodatskaya, E. & Kuzyakov, Y. Mechanisms of real and apparent priming effects and their dependence on soil microbial biomass and community structure: critical review. Biol. Fertil. Soils. 45, 115–131 (2008).
Nottingham, A. T., Turner, B. L., Chamberlain, P. M., Stott, A. W. & Tanner, E. V. J. Priming and microbial nutrient limitation in lowland tropical forest soils of contrasting fertility. Biogeochemistry. 111, 219–237 (2012).
Stewart, C. E., Moturi, P., Follett, R. F. & Halvorson, A. D. Lignin biochemistry and soil N determine crop residue decomposition and soil priming. Biogeochemistry. 124, 335–351 (2015).
Lonardo, D. P. Di et al. Priming of soil organic matter: chemical structure of added compounds is more important than the energy content. Soil Biol. Biochem. 108, 41–54 (2017).
Zwetsloot, M. J. et al. Prevalent root-derived phenolics drive shifts in microbial community composition and prime decomposition in forest soil. Soil Biol. Biochem. 145, 530–541 (2020).
Tao, X. et al. Winter warming in Alaska accelerates lignin decomposition contributed by Proteobacteria. Microbiome 8, 84 (2020).
Wutzler, T. & Reichstein, M. Priming and substrate quality interactions in soil organic matter models. Biogeosciences. 10, 2089–2103 (2013).
Guenet, B. et al. Impact of priming on global soil carbon stocks. Glob Chang Biol. 24, 1873–1883 (2018).
Schöning, I. & Kögel-Knabner, I. Chemical composition of young and old carbon pools throughout Cambisol and Luvisol profiles under forests. Soil Biol. Biochem. 38, 2411–2424 (2006).
Kleber, M. et al. Old and stable soil organic matter is not necessarily chemically recalcitrant: implications for modeling concepts and temperature sensitivity. Glob Chang Biol. 17, 1097–1107 (2011).
Northup, R. R., Dahlgren, R. A. & Yu, Z. Intraspecific variation of conifer phenolic concentration on a marine terrace soil acidity gradient; a new interpretation. Plant Soil. 171, 255–262 (1995).
Sanger, L. J., Cox, P., Splatt, P., Whelan, M. J. & Anderson, J. M. Variability in the quality of Pinus sylvestris needles and litter from sites with different soil characteristics: Lignin and phenylpropanoid signature. Soil Biol. Biochem. 28, 829–835 (1996).
Thevenot, M., Dignac, M. F. & Rumpel, C. Fate of lignins in soils: a review. Soil Biol. Biochem. 42, 1200–1211 (2010).
Zwetsloot, M. J. & Bauerle, T. L. Phenolic root exudate and tissue compounds vary widely among temperate forest tree species and have contrasting effects on soil microbial respiration. New Phytol. 218, 530–541 (2018).
Burges, N., Hurst, H. & Walkden, B. The phenolic constituents of humic acid and their relation to the lignin of the plant cover. Geochim. Cosmochim. Acta. 28, 1547–1554 (1964).
Kuiters, A. T. & Denneman, C. A. J. Water-soluble phenolic substances in soils under several coniferous and deciduous tree species. Soil Biol. Biochem. 19, 765–769 (1987).
Jalal, M. A. F. & Read, D. J. The organic acid composition of Calluna heathland soil with special reference to phyto- and fungitoxicity. Plant Soil. 70, 273–286 (1983).
Shindo, H., Ohta, S. & Kuwatsuka, S. Behavior of phenolic substances in the decaying process of plants: IX. Distribution of phenolic acids in soils of paddy fields and forests. Soil Sci. Plant Nutr. 24, 233–243 (1978).
Whitehead, D. C., Dibb, H. & Hartley, R. D. Extractant pH and the release of phenolic compounds from soils, plant roots and leaf litter. Soil Biol. Biochem. 13, 343–348 (1981).
Ed, V., Boyd, S. & Mokma, D. Extraction of phenolic compounds from a spodsol profile. Soil Sci. 140, 412–420 (1985).
Wang, Y. et al. Environmental behaviors of phenolic acids dominated their rhizodeposition in boreal poplar plantation forest soils. J. Soils Sediments. 16, 1858–1870 (2016).
Phillips, R. P. et al. Tree species and mycorrhizal associations influence the magnitude of rhizosphere effects. Ecology. 87, 1302–1313 (2006).
Blum, U. & Shafer, S. R. Microbial populations and phenolic acids in soil. Soil Biol. Biochem. 20, 793–800 (1988).
Shafer, S. R. & Blum, U. Influence of Phenolic acids on microbial populations in the rhizosphere of cucumber. J. Chem. Ecol. 17, 369–389 (1991).
Eilers, K. G., Lauber, C. L., Knight, R. & Fierer, N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 42, 896–903 (2010).
Morrissey, E. M. et al. Phylogenetic organization of bacterial activity. ISME J. 10, 2336–2340 (2016).
Huang P., Wang T., Wang M., Wu M., Hsu N. Retention of phenolic acids by noncrystalline hydroxy-aluminum and-iron compounds and clay minerals of soils. Soil Sci. 123, 213–219 (1977).
Cecchi, A. M., Koskinen, W. C., Cheng, H. H. & Haider, K. Sorption-desorption of phenolic acids as affected by soil properties. Biol. Fertil. Soils. 39, 235–242 (2004).
Shindo, H. & Kuwatsuka, S. Behavior of phenolic substances in the decaying process of plants: IV adsorption and movement of phenolic acids in soils. Soil Sci. Plant Nutr. 22, 23–33 (1976).
DeAngelis K. M. et al. Characterization of trapped lignin-degrading microbes in tropical forest soil. PLoS ONE. 6, e19306 (2011).
Pold G., Melillo J. M., DeAngelis K. M. Two decades of warming increases diversity of a potentially lignolytic bacterial community. Front Microbiol. 6, 480 (2015).
Wilhelm, R. C., Singh, R., Eltis, L. D. & Mohn, W. W. Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. 13, 413–429 (2019).
Folman, L. B., Klein Gunnewiek, P. J. A., Boddy, L., De & Boer, W. Impact of white-rot fungi on numbers and community composition of bacteria colonizing beech wood from forest soil. FEMS Microbiol. Ecol. 63, 181–191 (2008).
Valášková, V. et al. Phylogenetic composition and properties of bacteria coexisting with the fungus Hypholoma fasciculare in decaying wood. ISME J. 3, 1218–1221 (2009).
Mandal, S. M., Chakraborty, D. & Dey, S. Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal Behav. 5, 359–368 (2010).
Munoz Aguilar, M. et al. Chemotaxis of rhizobium leguminosarum biovar phaseoli towards flavonoid inducers of the symbiotic nodulation genes. J. Gen. Microbiol. 134, 2741–2746 (1988).
Morrissey, E. M. et al. Bacterial carbon use plasticity, phylogenetic diversity and the priming of soil organic matter. ISME J. 11, 1890–1899 (2017).
Fontaine, S., Mariotti, A. & Abbadie, L. The priming effect of organic matter: a question of microbial competition? Soil Biol. Biochem. 35, 837–843 (2003).
Liu, X. J. A. et al. The soil priming effect: consistent across ecosystems, elusive mechanisms. Soil Biol Biochem. 140, 107617 (2020).
Fanin, N., Alavoine, G. & Bertrand, I. Temporal dynamics of litter quality, soil properties and microbial strategies as main drivers of the priming effect. Geoderma [Internet]. 377, 114576 (2020).
Fuchs, G., Boll, M. & Heider, J. Microbial degradation of aromatic compounds - from one strategy to four. Nat. Rev. Microbiol. 9, 803–816 (2011).
Yang, Z. H. & Ji, G. D. Quantitative response relationships between degradation rates and functional genes during the degradation of beta-cypermethrin in soil. J. Hazard Mater. 299, 719–724 (2015).
Nishiyama, E., Ohtsubo, Y., Nagata, Y. & Tsuda, M. Identification of Burkholderia multivorans ATCC 17616 genes induced in soil environment by in vivo expression technology. Environ. Microbiol. 12, 2539–2558 (2010).
Wilhelm et al. Paraburkholderia madseniana sp. nov., a phenolic acid-degrading bacterium isolated from acidic forest soil. Int. J. Syst. Evol. Microbiol. 70, (2020). https://doi.org/10.1099/ijsem.0.004029.
Pallant, E. & Riha, S. J. Surface soil acidification under red pine and Norway spruce. Soil Sci. Soc. Am. J. 54, 1124–1130 (1990).
Fahey T. J. et al. Earthworm effects on the incorporation of litter C and N into soil organic matter in a sugar maple forest. Ecol. Appl. 23, 1185–1201 (2013).
Melvin, A. M. & Goodale, C. L. Tree species and earthworm effects on soil nutrient distribution and turnover in a northeastern United States common garden. Can. J. For. Res. 43, 180–187 (2013).
Suarez E. Invasion of Northern Hardwood Forests by Exotic Earthworm Communities in South-Central New York. Cornell; 2004.
Greweling T., Peech M. Chemical soil tests. Ithaca; 1960.
Griffiths, R. I., Whiteley, A. S., O’Donnell, A. G. & Bailey, M. J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66, 5488–5491 (2000).
Neufeld, J. D. et al. DNA stable-isotope probing. Nat. Protoc. 2, 860–866 (2007).
Wilhelm, R., Szeitz, A., Klassen, T. L. & Mohn, W. W. Sensitive, efficient quantitation of 13C-enriched nucleic acids via ultrahigh-performance liquid chromatography-tandem mass spectrometry for applications in stable isotope probing. Appl. Environ. Microbiol. 80, 7206–7211 (2014).
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
De Caceres, M. & Legendre, P. Associations between species and groups of sites: indices and statistical inference. Ecology, Ecology. 90, 3566–3574 (2009). http://sites.google.com/site/miqueldecaceres/.
Wilhelm, R. C., Niederberger, T. D., Greer, C. & Whyte, L. G. Microbial diversity of active layer and permafrost in an acidic wetland from the Canadian High Arctic. Can. J. Microbiol. 57, 303–315 (2011).
Ye, J. et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 13, 134 (2012).
López-Gutiérrez, J. C. et al. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Methods. 57, 399–407 (2004).
Markowitz, V. M. et al. IMG/M: A data management and analysis system for metagenomes. Nucleic Acids Res. 36(Suppl. 1), 534–538 (2008).
Martiny, A. C., Treseder, K. & Pusch, G. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 7, 830–838 (2013).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Callahan, B. J., McMurdie, P. J. & Holmes, S. P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643 (2017).
Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41(D1), 590–596 (2013).
Wickham, H. Elegant graphics for data analysis. Media. 35, 211 (2009).
McMurdie P. J., Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 8, e61217 (2013).
Oksanen J. et al. Vegan: community ecology package. R Packag. 2015;
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Price M. N., Dehal P. S., Arkin A. P. FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS ONE. 5, e9490 (2010).
Wilhelm R. C. et al. Paraburkholderia solitsugae sp. nov. and Paraburkholderia elongata sp. nov., phenolic acid-degrading bacteria isolated from forest soil and emended description of Paraburkholderia madseniana. Int. J. Syst. Evol. Microbiol. 70, 5093–5105 (2020).
Chain, P. S. G. et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. PNAS. 103, 15280–15287 (2006).
Mason-Jones, K. & Kuzyakov, Y. “Non-metabolizable” glucose analogue shines new light on priming mechanisms: Triggering of microbial metabolism. Soil Biol. Biochem. 107, 68–76 (2017).
Yuan, Y. et al. Exudate components exert different influences on microbially mediated C losses in simulated rhizosphere soils of a spruce plantation. Plant Soil. 419, 127–140 (2017).
Liu, X. J. A. et al. Labile carbon input determines the direction and magnitude of the priming effect. Appl. Soil Ecol. 109, 7–13 (2017).
Sugai, S. F. & Schimel, J. P. Decomposition and biomass incorporation of 14C-labeled glucose and phenolics in taiga forest floor: effect of substrate quality, successional state, and season. Soil Biol. Biochem. 25, 1379–1389 (1993).
Fontaine, S. et al. Fungi mediate long term sequestration of carbon and nitrogen in soil through their priming effect. Soil Biol. Biochem. 43, 86–96 (2011).
Zhu, Z. et al. Microbial stoichiometric flexibility regulates rice straw mineralization and its priming effect in paddy soil. Soil Biol. Biochem. 121, 67–76 (2018).
Roller, B. R. K., Stoddard, S. F. & Schmidt, T. M. Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat. Microbiol. 1, 1–7 (2016).
Smirnova, G. V. & Oktyabrsky, O. N. Relationship between Escherichia coli growth rate and bacterial susceptibility to ciprofloxacin. FEMS Microbiol. Lett. 365, 1–6 (2018).
Klappenbach, J. A., Dunbar, J. M. & Schmidt, T. M. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66, 1328–1333 (2000).
Méndez V., Agulló L., González M., Seeger M. The homogentisate and homoprotocatechuate central pathways are involved in 3- and 4-hydroxyphenylacetate degradation by Burkholderia xenovorans LB400. PLoS ONE. 6, e17583 (2011).
Johnson, G. R. & Olsen, R. H. Multiple pathways for toluene degradation in Burkholderia sp. strain JS150. Appl. Environ. Microbiol. 63, 4047–4052 (1997).
Andreolli, M. et al. Endophytic Burkholderia fungorum DBT1 can improve phytoremediation efficiency of polycyclic aromatic hydrocarbons. Chemosphere. 92, 688–694 (2013).
Somtrakoon, K. et al. Phenanthrene stimulates the degradation of pyrene and fluoranthene by Burkholderia sp. VUN10013. World J. Microbiol. Biotechnol. 24, 523–531 (2008).
Chain, P. S. G. et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. 103, 15280–15287 (2006).
Raj, A., Krishna Reddy, M. M. & Chandra, R. Identification of low molecular weight aromatic compounds by gas chromatography-mass spectrometry (GC-MS) from kraft lignin degradation by three Bacillus sp. Int. Biodeterior Biodegrad. 59, 292–296 (2007).
Shi, Y. et al. Biochemical investigation of kraft lignin degradation by Pandoraea sp. B-6 isolated from bamboo slips. Bioprocess Biosyst. Eng. 36, 1957–1965 (2013).
Moraes, E. C. et al. Lignolytic-consortium omics analyses reveal novel genomes and pathways involved in lignin modification and valorization. Biotechnol. Biofuels. 11, 1–16 (2018).
Coenye, T. et al. Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov., two new species isolated from the environment, animals and human clinical samples. Int. J. Syst. Evol. Microbiol. 51, 1099–1107 (2001).
Lim, Y. W., Baik, K. S., Han, S. K., Kim, S. B. & Bae, K. S. Burkholderia sordidicola sp. nov., isolated from the white-rot fungus Phanerochaete sordida. Int. J. Syst. Evol. Microbiol. 53, 1631–1636 (2003).
Herzog C. et al. Microbial succession on decomposing root litter in a drought-prone Scots pine forest. ISME J. 13, 2346–2362 (2019).
Yeoh Y. K. et al. Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat. Commun. 8, 215 (2017).
Zhalnina, K. et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 1–11 (2018).
Badri, D. V., Chaparro, J. M., Zhang, R., Shen, Q. & Vivanco, J. M. Application of natural blends of phytochemicals derived from the root exudates of arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 288, 4502–4512 (2013).
Sinsabaugh, R. L. Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 42, 391–404 (2010).
Henning, J. A. et al. Root bacterial endophytes alter plant phenotype, but not physiology. PeerJ. 2016, 1–20 (2016).
Caballero-Mellado, J., Martínez-Aguilar, L., Paredes-Valdez, G., & Estrada-de los Santos, P. Burkholderia unamae sp. nov., an N2-fixing rhizospheric and endophytic species. Int. J. Syst. Evol. Microbiol. 54, 1165–1172 (2004).
Martínez-Aguilar, L. et al. Burkholderia caballeronis sp. nov., a nitrogen fixing species isolated from tomato (Lycopersicon esculentum) with the ability to effectively nodulate Phaseolus vulgaris. Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol. 104, 1063–1071 (2013).
De Meyer, S. E. et al. Symbiotic and non-symbiotic Paraburkholderia isolated from South African Lebeckia ambigua root nodules and the description of Paraburkholderia fynbosensis sp. Nov. Int. J. Syst. Evol. Microbiol. 68, 2607–2614 (2018).
Peeters, C. et al. Phylogenomic study of Burkholderia glathei-like organisms, proposal of 13 novel Burkholderia species and emended descriptions of Burkholderia sordidicola, Burkholderia zhejiangensis, and Burkholderia grimmiae. Front Microbiol. 7, 1–19 (2016).
Vandamme, P. et al. Burkholderia humi sp. nov., Burkholderia choica sp. nov., Burkholderia telluris sp. nov., Burkholderia terrestris sp. nov. and Burkholderia udeis sp. nov.: Burkholderia glathei-like bacteria from soil and rhizosph. Int. J. Syst. Evol. Microbiol. 63(PART 12), 4707–4718 (2013).
Shiraishi, A., Matsushita, N. & Hougetsu, T. Nodulation in black locust by the Gammaproteobacteria Pseudomonas sp. and the Betaproteobacteria Burkholderia sp. Syst. Appl. Microbiol. 33, 269–274 (2010).
Thijs, S. et al. Exploring the rhizospheric and endophytic bacterial communities of Acer pseudoplatanus growing on a TNT-contaminated soil: towards the development of a rhizocompetent TNT-detoxifying plant growth promoting consortium. Plant Soil. 385, 15–36 (2014).
Mavengere, N. R., Ellis, A. G. & Le Roux, J. J. Burkholderia aspalathi sp. nov., isolated from root nodules of the South African legume Aspalathus abietina Thunb. Int. J. Syst. Evol. Microbiol. 64(PART 6), 1906–1912 (2014).
Blair, P. M. et al. Exploration of the biosynthetic potential of the populus microbiome. mSystems. 3, 1–17 (2018).
Peters, N. K. & Verma, D. P. S. Phenolic compounds as regulators of gene expression in plant-microbe interactions. Mol. Plant-Microbe Interact. 3, 4–8 (1990).
Kuzyakov, Y. & Blagodatskaya, E. Microbial hotspots and hot moments in soil: concept & review. Soil Biol. Biochem. 83, 184–199 (2015).
Acknowledgements
We acknowledge the intellectual contributions of Drs. Marie Zwetsloot and Taryn Bauerle for their foundational research on root exudates and soil priming. We thank Drs. Murray McBride, Dominic Woolf and Timothy Fahey for their discussion of soil chemical, biogeochemistry and forest soil ecology, and Patricia Brenchley and Derrick Barret for their effective management of project resources. This work was supported by McIntire Stennis under NYC-189502 from the USDA National Institute of Food and Agriculture, and by the U.S. Department of Energy, Office of Biological & Environmental Research Genomic Science Program under award number DE-SC0016364. We would like to highlight the contributions of the late Dr. Eugene Madsen who initiated this research project prior to his untimely passing. May his life’s work be an inspiration to future environmental microbiologists.
Author information
Authors and Affiliations
Contributions
R.C.W. performed all data analysis, research and writing, as well as designing the priming experiment and assisting field work. C.M.D. performed all field experiments, methods development, including pobA primer design, and assisted with the priming experiment. J.P.S. identified the pobA paralogs and provided valuable feedback in preparation of the manuscript. E.L.M. conceived of the central aims of the study, selected the field site and designed and performed all field experiments. D.H.B. guided all research efforts and contributed significantly to designing the priming experiment, interpreting data and writing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wilhelm, R.C., DeRito, C.M., Shapleigh, J.P. et al. Phenolic acid-degrading Paraburkholderia prime decomposition in forest soil. ISME COMMUN. 1, 4 (2021). https://doi.org/10.1038/s43705-021-00009-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43705-021-00009-z