Abstract
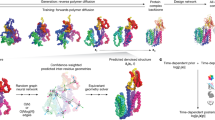
The generation of de novo protein structures with predefined functions and properties remains a challenging problem in protein design. Diffusion models, also known as score-based generative models (SGMs), have recently exhibited astounding empirical performance in image synthesis. Here we use image-based representations of protein structure to develop ProteinSGM, a score-based generative model that produces realistic de novo proteins. Through unconditional generation, we show that ProteinSGM can generate native-like protein structures, surpassing the performance of previously reported generative models. We experimentally validate some de novo designs and observe secondary structure compositions consistent with generated backbones. Finally, we apply conditional generation to de novo protein design by formulating it as an image inpainting problem, allowing precise and modular design of protein structure.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The training data used for this work is the CATH 4.3 non-redundant S40 dataset, which can be found here: http://download.cathdb.info/cath/releases/all-releases/v4_3_0/non-redundant-data-sets/cath-dataset-nonredundant-S40-v4_3_0.pdb.tgz. The dataset was filtered for structure lengths of between 40 and 128 (see Methods)—the exact CATH IDs used for the training/test splits can be found in the repository linked below. Source Data are provided with this paper.
Code availability
The codebase used for this work is available at https://gitlab.com/mjslee0921/proteinsgm and the Zenodo repository41.
References
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Strokach, A., Becerra, D., Corbi-Verge, C., Perez-Riba, A. & Kim, P. M. Fast and flexible protein design using deep graph neural networks. Cell Syst. 11, 402–411.e4 (2020).
Hsu, C. et al. Learning inverse folding from millions of predicted structures. In International Conference on Machine Learning 8946–8970 (PMLR, 2022).
Dauparas, J. et al. Robust deep learning based protein sequence design using ProteinMPNN. Science 378, 49–56 (2022).
Andreeva, A., Kulesha, E., Gough, J. & Murzin, A. G. The SCOP database in 2020: expanded classification of representative family and superfamily domains of known protein structures. Nucl. Acids Res. 48, D376–D382 (2020).
Sillitoe, I. et al. CATH: increased structural coverage of functional space. Nucl. Acids Res. 49, D266–D273 (2021).
Wang, J. et al. Scaffolding protein functional sites using deep learning. Science 377, 387–394 (2022).
Ho, J., Jain, A. & Abbeel, P. Denoising diffusion probabilistic models. In Advances in Neural Information Processing Systems Vol. 33, 6840–6851 (Curran Associates, 2020).
Song, Y. & Ermon, S. Generative modeling by estimating gradients of the data distribution. In Advances in Neural Information Processing Systems Vol. 32 (Curran Associates, 2019).
Dhariwal, P. & Nichol, A. Diffusion models beat gans on image synthesis. In Advances in Neural Information Processing Systems Vol. 34, 8780–8794 (Curran Associates, 2021).
Kong, Z., Ping, W., Huang, J., Zhao, K. & Catanzaro, B. Diffwave: a versatile diffusion model for audio synthesis. In International Conference of Learning Representations (ICLR, 2021).
Ramesh, A., Dhariwal, P., Nichol, A., Chu, C. & Chen, M. Hierarchical text-conditional image generation with CLIP latents. Preprint at http://arxiv.org/abs/2204.06125 (2022).
Niu, C. et al. Permutation invariant graph generation via score-based generative modeling. In International Conference on Artificial Intelligence and Statistics 4474–4484 (PMLR, 2020).
Jo, J., Lee, S. & Hwang, S. J. Score-based generative modeling of graphs via the system of stochastic differential equations. In International Conference on Machine Learning 10362–10383 (PMLR, 2022).
Hoogeboom, E., Satorras, V. G., Vignac, C. & Welling, M. Equivariant diffusion for molecule generation in 3d. In International Conference on Machine Learning 8867–8887 (PMLR, 2022).
Song, Y. et al. Score-based generative modeling through stochastic differential equations. In International Conference of Learning Representations (ICLR, 2020).
Anand, N. & Achim, T. Protein structure and sequence generation with equivariant denoising diffusion probabilistic models. Preprint at http://arxiv.org/abs/2205.15019 (2022).
Trippe, B. L. et al. Diffusion probabilistic modeling of protein backbones in 3D for the motif-scaffolding problem. In International Conference of Learning Representations (ICLR, 2022).
Wu, K. E. et al. Protein structure generation via folding diffusion. Preprint at http://arxiv.org/abs/2209.15611 (2022).
Watson, J. L. et al. Broadly applicable and accurate protein design by integrating structure prediction networks and diffusion generative models. Preprint at http://biorxiv.org/lookup/doi/10.1101/2022.12.09.519842 (2022).
Ingraham, J. et al. Illuminating protein space with a programmable generative model. Preprint at http://biorxiv.org/lookup/doi/10.1101/2022.12.01.518682 (2022).
Wu, R. et al. High-resolution de novo structure prediction from primary sequence. Preprint at http://biorxiv.org/lookup/doi/10.1101/2022.07.21.500999 (2022).
Yang, J. et al. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl Acad. Sci. USA 117, 1496–1503 (2020).
Lin, Z., Sercu, T., LeCun, Y. & Rives, A. Deep generative models create new and diverse protein structures. Machine Learning in Structural Biology (NeurIPS, 2021).
Zhang, Y. & Skolnick, J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucl. Acids Res. 33, 2302–2309 (2005).
Xu, J. & Zhang, Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 26, 889–895 (2010).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Micsonai, András et al. BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucl. Acids Res. 46, W315–W322 (2018).
Greenfield, N. J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protocols 1, 2876–2890 (2006).
Rombach, R., Blattmann, A., Lorenz, D., Esser, P. & Ommer, B. High-resolution image synthesis with latent diffusion models. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 10684–10695 (IEEE, 2022).
Van Rossum, G. & Drake, F. L. Python 3 Reference Manual (CreateSpace, 2009).
Harris, C. R. et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Paszke, A. et al. Pytorch: an imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems Vol. 32, 8024–8035 (Curran Associates, 2019); http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf
Cock, P. J. A. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009).
Kunzmann, P. & Hamacher, K. Biotite: a unifying open source computational biology framework in python. BMC Bioinform. 19, 346 (2018).
Vincent, P. A connection between score matching and denoising autoencoders. Neural Comput. 23, 1661–1674 (2011).
Lin, G., Milan, A., Shen, C. & Reid, I. Refinenet: multi-path refinement networks for high-resolution semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 1925–1934 (IEEE, 2017).
Alford, R. F. et al. The rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theory Comput. 13, 3031–3048 (2017).
Chaudhury, S., Lyskov, S. & Gray, J. J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010).
Lee, J. S., Kim, J. & Kim, P. M. Proteinsgm Codebase (Zenodo, 2023); https://doi.org/10.5281/zenodo.7755375
Acknowledgements
We acknowledge the CIHR Project Grant (grant no. PJT-153279) and NSERC Discovery Grant (grant no. RGPIN-2017-064) for funding. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We also thank the Digital Research Alliance of Canada for computing resources.
Author information
Authors and Affiliations
Contributions
J.S.L. and P.M.K. conceptualized the work, J.S.L. developed the computational results, J.S.K. performed the experimental validation and J.S.L., J.S.K., and P.M.K. wrote the manuscript. P.M.K. supervised the work and acquired funding.
Corresponding author
Ethics declarations
Competing interests
P.M.K. is a co-founder and consultant to multiple companies, including Resolute Bio, Oracle Therapeutics and Navega Therapeutics and serves on the scientific advisory board of ProteinQure. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Andrea Ventura and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, and Tables 1 and 2.
Source data
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lee, J.S., Kim, J. & Kim, P.M. Score-based generative modeling for de novo protein design. Nat Comput Sci 3, 382–392 (2023). https://doi.org/10.1038/s43588-023-00440-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-023-00440-3
This article is cited by
-
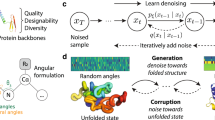
Protein structure generation via folding diffusion
Nature Communications (2024)
-
Opportunities and challenges in design and optimization of protein function
Nature Reviews Molecular Cell Biology (2024)
-
Sparks of function by de novo protein design
Nature Biotechnology (2024)
-
Artificial intelligence and illusions of understanding in scientific research
Nature (2024)
-
From noise to protein with image models
Nature Computational Science (2023)