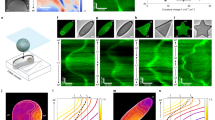
Abstract
How does breaking the symmetry of an equation alter the symmetry of its solutions? Here, we systematically examine how reducing underlying symmetries from spherical to axisymmetric influences the dynamics of an archetypal model of cell polarization, a key process of biological spatial self-organization. Cell polarization is characterized by nonlinear and non-local dynamics, but we overcome the theory challenges these traits pose by introducing a broadly applicable numerical scheme allowing us to efficiently study continuum models in a wide range of geometries. Guided by numerical results, we discover a dynamical hierarchy of timescales that allows us to reduce relaxation to a purely geometric problem of area-preserving geodesic curvature flow. Through application of variational results, we analytically construct steady states on a number of biologically relevant shapes. In doing so, we reveal non-trivial solutions for symmetry breaking.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Source data for all quantitative results are available with this manuscript and on Zenodo50.
Change history
17 October 2022
A Correction to this paper has been published: https://doi.org/10.1038/s43588-022-00345-7
References
Heath, T. L. et al. The Thirteen Books of Euclid’s Elements (Courier, 1956).
Gross, P. et al. Guiding self-organized pattern formation in cell polarity establishment. Nat. Phys. 15, 293–300 (2019).
Mietke, A., Jemseena, V., Kumar, K. V., Sbalzarini, I. F. & Jülicher, F. Minimal model of cellular symmetry breaking. Phys. Rev. Lett. 123, 188101 (2019).
Brauns, F. et al. Bulk–surface coupling identifies the mechanistic connection between Min-protein patterns in vivo and in vitro. Nat. Commun. 12, 3312 (2021).
Zhu, M. et al. Developmental clock and mechanism of de novo polarization of the mouse embryo. Science 370, eabd2703 (2020).
Chen, W., Nie, Q., Yi, T.-M. & Chou, C.-S. Modelling of yeast mating reveals robustness strategies for cell–cell interactions. PLoS Comput. Biol. 12, e1004988 (2016).
Rappel, W.-J. & Edelstein-Keshet, L. Mechanisms of cell polarization. Curr. Opin. Syst. Biol. 3, 43–53 (2017).
Tostevin, F., Wigbers, M., Søgaard-Andersen, L. & Gerland, U. Four different mechanisms for switching cell polarity. PLoS Comput. Biol. 17, e1008587 (2021).
Goryachev, A. B. & Leda, M. Many roads to symmetry breaking: molecular mechanisms and theoretical models of yeast cell polarity. Mol. Biol. Cell 28, 370–380 (2017).
Thalmeier, D., Halatek, J. & Frey, E. Geometry-induced protein pattern formation. Proc. Natl Acad. Sci. USA 113, 548–553 (2016).
Diegmiller, R., Montanelli, H., Muratov, C. B. & Shvartsman, S. Y. Spherical caps in cell polarization. Biophys. J. 115, 26–30 (2018).
Bäcker, J. P. & Röger, M. Analysis and asymptotic reduction of a bulk-surface reaction-diffusion model of Gierer-Meinhardt type. Commun. Pure Appl. Math. 21, 1139 (2022).
Gamba, A., Kolokolov, I., Lebedev, V. & Ortenzi, G. Universal features of cell polarization processes. J. Stat. Mech. Theory Exp. 2009, P02019 (2009).
Cusseddu, D., Edelstein-Keshet, L., Mackenzie, J. A., Portet, S. & Madzvamuse, A. A coupled bulk–surface model for cell polarisation. J. Theor. Biol. 481, 119–135 (2019).
Geßele, R., Halatek, J., Würthner, L. & Frey, E. Geometric cues stabilise long-axis polarisation of PAR protein patterns in C. elegans. Nat. Commun. 11, 539 (2020).
Rätz, A. & Röger, M. Symmetry breaking in a bulk–surface reaction–diffusion model for signalling networks. Nonlinearity 27, 1805 (2014).
Trogdon, M. et al. The effect of cell geometry on polarization in budding yeast. PLoS Comput. Biol. 14, e1006241 (2018).
Golubitsky, M., Stewart, I. & Schaeffer, D. G. Singularities and Groups in Bifurcation Theory: Volume II (Applied Mathematical Sciences Vol. 69, Springer, 2012).
Merilees, P. E. The pseudospectral approximation applied to the shallow water equations on a sphere. Atmosphere 11, 13–20 (1973).
Orszag, S. A. Fourier series on spheres. Mon. Weather Rev. 102, 56–75 (1974).
Townsend, A., Wilber, H. & Wright, G. B. Computing with functions in spherical and polar geometries I. The sphere. SIAM J. Sci. Comput. 38, C403–C425 (2016).
Goryachev, A. B. & Pokhilko, A. V. Dynamics of cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 582, 1437–1443 (2008).
Otsuji, M. et al. A mass conserved reaction–diffusion system captures properties of cell polarity. PLoS Comput. Biol. 3, e108 (2007).
Mori, Y., Jilkine, A. & Edelstein-Keshet, L. Wave-pinning and cell polarity from a bistable reaction–diffusion system. Biophys. J. 94, 3684–3697 (2008).
Sharma, V. & Morgan, J. Global existence of solutions to reaction–diffusion systems with mass transport type boundary conditions. SIAM J. Math. Anal. 48, 4202–4240 (2016).
Fortunato, D. Spectral methods for reaction–diffusion on axisymmetric surfaces. GitHub https://github.com/danfortunato/surface-diffusion (2022).
Fortunato, D. danfortunato/surface-diffusion (v1.0.0). Zenodo https://doi.org/10.5281/zenodo.6762738 (2022).
Ihrig, E. & Golubitsky, M. Pattern selection with O(3) symmetry. Physica D 13, 1–33 (1984).
Henry, M., Hilhorst, D. & Muratov, C. B. A multiple scale pattern formation cascade in reaction–diffusion systems of activator–inhibitor type. Interfaces Free Bound. 20, 297–336 (2018).
Camley, B. A., Zhao, Y., Li, B., Levine, H. & Rappel, W.-J. Crawling and turning in a minimal reaction–diffusion cell motility model: coupling cell shape and biochemistry. Phys. Rev. E 95, 012401 (2017).
Vanderlei, B., Feng, J. J. & Edelstein-Keshet, L. A computational model of cell polarization and motility coupling mechanics and biochemistry. Multiscale Model. Simul. 9, 1420–1443 (2011).
Brauns, F., Weyer, H., Halatek, J., Yoon, J. & Frey, E. Wavelength selection by interrupted coarsening in reaction–diffusion systems. Phys. Rev. Lett. 126, 104101 (2021).
Ritoré, M. & Sinestrari, C. Mean Curvature Flow and Isoperimetric Inequalities (Springer, 2010).
Rubinstein, J. & Sternberg, P. Nonlocal reaction–diffusion equations and nucleation. IMA J. Appl. Math. 48, 249–264 (1992).
Ros, A. The isoperimetric problem. Glob. Theory Minim. Surf. 2, 175–209 (2001).
Morgan, F. & Johnson, D. L. Some sharp isoperimetric theorems for Riemannian manifolds. Indiana Univ. Math. J. 49, 1017–1041 (2000).
Ritoré, M. Constant geodesic curvature curves and isoperimetric domains in rotationally symmetric surfaces. Commun. Anal. Geom. 9, 1093–1138 (2001).
Fonda, P., Rinaldin, M., Kraft, D. J. & Giomi, L. Interface geometry of binary mixtures on curved substrates. Phys. Rev. E 98, 032801 (2018).
Wigbers, M. C. et al. A hierarchy of protein patterns robustly decodes cell shape information. Nat. Phys. 17, 578–584 (2021).
Zhang, Z., Zwick, S., Loew, E., Grimley, J. S. & Ramanathan, S. Mouse embryo geometry drives formation of robust signaling gradients through receptor localization. Nat. Commun. 10, 4516 (2019).
Tan, T. H. et al. Topological turbulence in the membrane of a living cell. Nat. Phys. 16, 657–662 (2020).
Deneke, V. E., Melbinger, A., Vergassola, M. & Di Talia, S. Waves of cdk1 activity in S phase synchronize the cell cycle in Drosophila embryos. Dev. Cell 38, 399–412 (2016).
Liu, J. et al. Topological braiding and virtual particles on the cell membrane. Proc. Natl Acad. Sci. USA 118, e2104191118 (2021).
Miller, P. W., Stoop, N. & Dunkel, J. Geometry of wave propagation on active deformable surfaces. Phys. Rev. Lett. 120, 268001 (2018).
Baker, R. E. & Maini, P. A mechanism for morphogen-controlled domain growth. J. Math. Biol. 54, 597–622 (2007).
Gomez, D., Iyaniwura, S., Paquin-Lefebvre, F. & Ward, M. Pattern forming systems coupling linear bulk diffusion to dynamically active membranes or cells. Philos. Trans. R. Soc. A 379, 20200276 (2021).
Kirk, B. S., Peterson, J. W., Stogner, R. H. & Carey, G. F. libMesh: a C++ library for parallel adaptive mesh refinement/coarsening simulations. Eng. Comput. 22, 237–254 (2006).
Calvo, M., de Frutos, J. & Novo, J. Linearly implicit Runge–Kutta methods for advection–reaction–diffusion equations. Appl. Numer. Math. 37, 535–549 (2001).
Miller, P. W. & Dunkel, J. Gait-optimized locomotion of wave-driven soft sheets. Soft Matter 16, 3991–3999 (2020).
Miller, P. Source data for ‘Forced and spontaneous symmetry breaking in cell polarization’. Zenodo https://doi.org/10.5281/zenodo.6774314 (2022).
Acknowledgements
We thank Rocky Diegmiller, Boris Slepchenko, Martin Golubistky and Matteo Novaga for helpful discussions, and Lucy Reading-Ikkanda for assistance with graphical design. This work was supported by NIH grant R01 GM134204 to S.S.
Author information
Authors and Affiliations
Contributions
S.S., L.G. and C.M. designed the research. P.W.M. performed analytical and numerical studies. D.F. developed the numerical method and software. All authors discussed the results and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Johannes Borgqvist, Anotida Madzvamuse and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Ananya Rastogi, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Discussion, Figs. 1–4 and Table 1.
Supplementary Video 1
Simulation of cell polarization on a variety of surfaces.
Supplementary Video 2
Different initial conditions on an egg-shaped surface.
Source data
Source Data Fig. 2
Numerical source data.
Source Data Fig. 3
Numerical source data.
Source Data Fig. 4
Numerical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Miller, P.W., Fortunato, D., Muratov, C. et al. Forced and spontaneous symmetry breaking in cell polarization. Nat Comput Sci 2, 504–511 (2022). https://doi.org/10.1038/s43588-022-00295-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-022-00295-0
This article is cited by
-
A double Fourier sphere method for d-dimensional manifolds
Sampling Theory, Signal Processing, and Data Analysis (2023)
-
Geometry and symmetry-breaking in cell polarity
Nature Computational Science (2022)