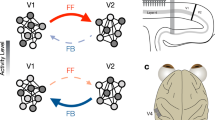
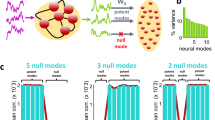
Abstract
Technological advances now allow us to record from large populations of neurons across multiple brain areas. These recordings may illuminate how communication between areas contributes to brain function, yet a substantial barrier remains: how do we disentangle the concurrent, bidirectional flow of signals between populations of neurons? We propose here a dimensionality reduction framework, delayed latents across groups (DLAG), that disentangles signals relayed in each direction, identifies how these signals are represented by each population and characterizes how they evolve within and across trials. We demonstrate that DLAG performs well on synthetic datasets similar in scale to current neurophysiological recordings. Then we study simultaneously recorded populations in primate visual areas V1 and V2, where DLAG reveals signatures of bidirectional yet selective communication. Our framework lays a foundation for dissecting the intricate flow of signals across populations of neurons, and how this signalling contributes to cortical computation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
V1–V2 data are available at the CRCNS data sharing website at https://doi.org/10.6080/K0B27SHN (ref. 70). Naturalistic texture images are available on the Multiband Texture Database at http://multibandtexture.recherche.usherbrooke.ca/original_brodatz.html and on the Salzburg Texture Image Database at https://wavelab.at/sources/STex. Source Data for Figs. 3–6 are available for this Article.
Code availability
A MATLAB implementation of DLAG is available on GitHub at https://github.com/egokcen/DLAG and on Zenodo at https://doi.org/10.5281/zenodo.6654831 (ref. 71).
References
Ahrens, M. B. et al. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 485, 471–477 (2012).
Yang, W. & Yuste, R. In vivo imaging of neural activity. Nat. Methods 14, 349–359 (2017).
Jun, J. J. et al. Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236 (2017).
Steinmetz, N. A., Zatka-Haas, P., Carandini, M. & Harris, K. D. Distributed coding of choice, action and engagement across the mouse brain. Nature 576, 266–273 (2019).
Kohn, A. et al. Principles of corticocortical communication: proposed schemes and design considerations. Trends Neurosci. 43, 725–737 (2020).
Lamme, V. A., Supèr, H. & Spekreijse, H. Feedforward, horizontal, and feedback processing in the visual cortex. Curr. Opin. Neurobiol. 8, 529–535 (1998).
Angelucci, A. & Bressloff, P. C. Contribution of feedforward, lateral and feedback connections to the classical receptive field center and extra-classical receptive field surround of primate V1 neurons. Prog. Brain Res. 154, 93–120 (2006).
Gilbert, C. D. & Li, W. Top-down influences on visual processing. Nat. Rev. Neurosci. 14, 350–363 (2013).
Harris, K. D. & Mrsic-Flogel, T. D. Cortical connectivity and sensory coding. Nature 503, 51–58 (2013).
Miller, E. K., Lundqvist, M. & Bastos, A. M. Working Memory 2.0. Neuron 100, 463–475 (2018).
Shadmehr, R. & Krakauer, J. W. A computational neuroanatomy for motor control. Exp. Brain Res. 185, 359–381 (2008).
Keemink, S. W. & Machens, C. K. Decoding and encoding (de)mixed population responses. Curr. Opin. Neurobiol. 58, 112–121 (2019).
Schmolesky, M. T. et al. Signal timing across the macaque visual system. J. Neurophysiol. 79, 3272–3278 (1998).
Hernández, A. et al. Decoding a perceptual decision process across cortex. Neuron 66, 300–314 (2010).
Siegel, M., Buschman, T. J. & Miller, E. K. Cortical information flow during flexible sensorimotor decisions. Science 348, 1352–1355 (2015).
Supèr, H., Spekreijse, H. & Lamme, V. A. F. Two distinct modes of sensory processing observed in monkey primary visual cortex (V1). Nat. Neurosci. 4, 304–310 (2001).
Pooresmaeili, A., Poort, J. & Roelfsema, P. R. Simultaneous selection by object-based attention in visual and frontal cortex. Proc. Natl Acad. Sci. USA 111, 6467–6472 (2014).
Chen, M. et al. Incremental integration of global contours through interplay between visual cortical areas. Neuron 82, 682–694 (2014).
Schwiedrzik, C. M. & Freiwald, W. A. High-level prediction signals in a low-level area of the macaque face-processing hierarchy. Neuron 96, 89–97 (2017).
Issa, E. B., Cadieu, C. F. & DiCarlo, J. J. Neural dynamics at successive stages of the ventral visual stream are consistent with hierarchical error signals. eLife 7, e42870 (2018).
Reid, R. C. & Alonso, J. M. Specificity of monosynaptic connections from thalamus to visual cortex. Nature 378, 281–284 (1995).
Roe, A. W. & Ts’o, D. Y. Specificity of color connectivity between primate V1 and V2. J. Neurophysiol. 82, 2719–2730 (1999).
Nowak, L. G., Munk, M., James, A. C., Girard, P. & Bullier, J. Cross-correlation study of the temporal interactions between areas V1 and V2 of the macaque monkey. J. Neurophysiol. 81, 1057–1074 (1999).
Jia, X., Tanabe, S. & Kohn, A. Gamma and the coordination of spiking activity in early visual cortex. Neuron 77, 762–774 (2013).
Oemisch, M., Westendorff, S., Everling, S. & Womelsdorf, T. Interareal spike-train correlations of anterior cingulate and dorsal prefrontal cortex during attention shifts. J. Neurosci. 35, 13076–13089 (2015).
Zandvakili, A. & Kohn, A. Coordinated neuronal activity enhances corticocortical communication. Neuron 87, 827–839 (2015).
Campo, A. T. et al. Feed-forward information and zero-lag synchronization in the sensory thalamocortical circuit are modulated during stimulus perception. Proc. Natl Acad. Sci. USA 116, 7513–7522 (2019).
Gregoriou, G. G., Gotts, S. J., Zhou, H. & Desimone, R. High-frequency, long-range coupling between prefrontal and visual cortex during attention. Science 324, 1207–1210 (2009).
Salazar, R. F., Dotson, N. M., Bressler, S. L. & Gray, C. M. Content-specific fronto-parietal synchronization during visual working memory. Science 338, 1097–1100 (2012).
van Kerkoerle, T. et al. Alpha and gamma oscillations characterize feedback and feedforward processing in monkey visual cortex. Proc. Natl Acad. Sci. USA 111, 14332–14341 (2014).
Bastos, A. M., Vezoli, J. & Fries, P. Communication through coherence with inter-areal delays. Curr. Opin. Neurobiol. 31, 173–180 (2015).
Semedo, J. D., Gokcen, E., Machens, C. K., Kohn, A. & Yu, B. M. Statistical methods for dissecting interactions between brain areas. Curr. Opin. Neurobiol. 65, 59–69 (2020).
Kang, B. & Druckmann, S. Approaches to inferring multi-regional interactions from simultaneous population recordings. Curr. Opin. Neurobiol. 65, 108–119 (2020).
Keeley, S. L., Zoltowski, D. M., Aoi, M. C. & Pillow, J. W. Modeling statistical dependencies in multi-region spike train data. Curr. Opin. Neurobiol. 65, 194–202 (2020).
Kaufman, M. T., Churchland, M. M., Ryu, S. I. & Shenoy, K. V. Cortical activity in the null space: permitting preparation without movement. Nat. Neurosci. 17, 440–448 (2014).
Semedo, J. D., Zandvakili, A., Machens, C. K., Yu, B. M. & Kohn, A. Cortical areas interact through a communication subspace. Neuron 102, 249–259 (2019).
Perich, M. G., Gallego, J. A. & Miller, L. E. A neural population mechanism for rapid learning. Neuron 100, 964–976 (2018).
Srinath, R., Ruff, D. A. & Cohen, M. R. Attention improves information flow between neuronal populations without changing the communication subspace. Curr. Biol. 31, 5299–5313 (2021).
Veuthey, T. L., Derosier, K., Kondapavulur, S. & Ganguly, K. Single-trial cross-area neural population dynamics during long-term skill learning. Nat. Commun. 11, 4057 (2020).
Chen, G., Kang, B., Lindsey, J., Druckmann, S. & Li, N. Modularity and robustness of frontal cortical networks. Cell 184, 3717–3730 (2021).
Semedo, J. D. et al. Feedforward and feedback interactions between visual cortical areas use different population activity patterns. Nat. Commun. 13, 1099 (2022).
Bach, F. R. & Jordan, M. I. A Probabilistic Interpretation of Canonical Correlation Analysis Technical Report 688 (Department of Statistics, University of California, Berkeley, 2005).
Archambeau, C. & Bach, F. Sparse probabilistic projections. Adv. Neural Inf. Process. Syst. 21, 73–80 (2008).
Yu, B. M. et al. Gaussian-process factor analysis for low-dimensional single-trial analysis of neural population activity. J. Neurophysiol. 102, 614–635 (2009).
Lakshmanan, K. C., Sadtler, P. T., Tyler-Kabara, E. C., Batista, A. P. & Yu, B. M. Extracting low-dimensional latent structure from time series in the presence of delays. Neural Comput. 27, 1825–1856 (2015).
Felleman, D. J. & Van Essen, D. C. Distributed hierarchical processing in the primate cerebral cortex. Cereb. Cortex 1, 1–47 (1991).
Markov, N. T. et al. Cortical high-density counterstream architectures. Science 342, 1238406 (2013).
Smith, M. A., Kohn, A. & Movshon, J. A. Glass pattern responses in macaque V2 neurons. J. Vision 7, 5 (2007).
Murray, J. D. et al. A hierarchy of intrinsic timescales across primate cortex. Nat. Neurosci. 17, 1661–1663 (2014).
Runyan, C. A., Piasini, E., Panzeri, S. & Harvey, C. D. Distinct timescales of population coding across cortex. Nature 548, 92–96 (2017).
Siegle, J. H. et al. Survey of spiking in the mouse visual system reveals functional hierarchy. Nature 592, 86–92 (2021).
Zhuang, X., Yang, Z. & Cordes, D. A technical review of canonical correlation analysis for neuroscience applications. Hum. Brain Mapp. 41, 3807–3833 (2020).
Kamiński, M., Ding, M., Truccolo, W. A. & Bressler, S. L. Evaluating causal relations in neural systems: Granger causality, directed transfer function and statistical assessment of significance. Biol. Cybern. 85, 145–157 (2001).
Quinn, C. J., Coleman, T. P., Kiyavash, N. & Hatsopoulos, N. G. Estimating the directed information to infer causal relationships in ensemble neural spike train recordings. J. Comput. Neurosci. 30, 17–44 (2011).
Kim, S., Putrino, D., Ghosh, S. & Brown, E. N. A Granger causality measure for point process models of ensemble neural spiking activity. PLoS Comput. Biol. 7, e1001110 (2011).
Pillow, J. W. et al. Spatio-temporal correlations and visual signalling in a complete neuronal population. Nature 454, 995–999 (2008).
Truccolo, W., Hochberg, L. R. & Donoghue, J. P. Collective dynamics in human and monkey sensorimotor cortex: predicting single neuron spikes. Nat. Neurosci. 13, 105–111 (2010).
Perich, M. G. et al. Inferring brain-wide interactions using data-constrained recurrent neural network models. Preprint at https://doi.org/10.1101/2020.12.18.423348 (2021).
Rodu, J., Klein, N., Brincat, S. L., Miller, E. K. & Kass, R. E. Detecting multivariate cross-correlation between brain regions. J. Neurophysiol. 120, 1962–1972 (2018).
Bong, H. et al. Latent dynamic factor analysis of high-dimensional neural recordings. Adv. Neural Inf. Process. Syst. 33, 16446–16456 (2020).
Keeley, S., Aoi, M., Yu, Y., Smith, S. & Pillow, J. W. Identifying signal and noise structure in neural population activity with Gaussian process factor models. Adv. Neural Inf. Process. Syst. 33, 13795–13805 (2020).
Semedo, J., Zandvakili, A., Kohn, A., Machens, C. K. & Yu, B. M. Extracting latent structure from multiple interacting neural populations. Adv. Neural Inf. Process. Syst. 27, 2942–2950 (2014).
Glaser, J., Whiteway, M., Cunningham, J. P., Paninski, L. & Linderman, S. Recurrent switching dynamical systems models for multiple interacting neural populations. Adv. Neural Inf. Process. Syst. 33, 14867–14878 (2020).
Reid, A. T. et al. Advancing functional connectivity research from association to causation. Nat. Neurosci. 22, 1751–1760 (2019).
Rasmussen, C. E. & Williams, C. K. I. Gaussian Processes for Machine Learning (MIT Press, 2006).
Golub, G. H. & Van Loan, C. F. Matrix Computations 4th edn (Johns Hopkins Univ. Press, 2013).
Smith, M. A. & Kohn, A. Spatial and temporal scales of neuronal correlation in primary visual cortex. J. Neurosci. 28, 12591–12603 (2008).
Cowley, B. R. et al. Slow drift of neural activity as a signature of impulsivity in macaque visual and prefrontal cortex. Neuron 108, 551–567 (2020).
Pei, F. et al. Neural Latents Benchmark ’21: evaluating latent variable models of neural population activity. Preprint at https://doi.org/10.48550/arXiv.2109.04463 (2022).
Zandvakili, A. & Kohn, A. Simultaneous V1–V2 neuronal population recordings in anesthetized macaque monkeys. CRCNS https://doi.org/10.6080/K0B27SHN (2019).
Gokcen, E. egokcen/DLAG: v1.0.0. Zenodo https://doi.org/10.5281/zenodo.6654831 (2022).
Acknowledgements
This work was supported by the Dowd Fellowship (E.G.), Simons Collaboration on the Global Brain 542999 (A.K.), 543009 (C.K.M.), 543065 (B.M.Y.), 364994 (A.K., B.M.Y.), NIH R01 EY028626 (A.K.), NIH U01 NS094288 (C.K.M.), NIH R01 HD071686 (B.M.Y.), NIH CRCNS R01 NS105318 (B.M.Y.), NSF NCS BCS 1533672 and 1734916 (B.M.Y.), NIH CRCNS R01 MH118929 (B.M.Y.) and NIH R01 EB026953 (B.M.Y.).
Author information
Authors and Affiliations
Contributions
E.G., A.I.J., J.D.S., A.K., C.K.M. and B.M.Y. designed the analyses. E.G. derived and implemented DLAG, and performed all analyses. A.I.J., A.Z. and A.K. designed and performed the experiments. E.G., A.I.J., A.K., C.K.M. and B.M.Y. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Matthew Kaufman, Stephen Keeley, Stefano Recanatesi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Ananya Rastogi, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–17, Note and Discussion.
Source data
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 5
Statistical Source Data.
Source Data Fig. 6
Statistical Source Data.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gokcen, E., Jasper, A.I., Semedo, J.D. et al. Disentangling the flow of signals between populations of neurons. Nat Comput Sci 2, 512–525 (2022). https://doi.org/10.1038/s43588-022-00282-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-022-00282-5
This article is cited by
-
Multiregion neuronal activity: the forest and the trees
Nature Reviews Neuroscience (2022)
-
Untangling network information flow
Nature Computational Science (2022)