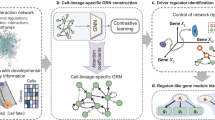
Abstract
Gene regulatory networks (GRNs) encode the complex molecular interactions that govern cell identity. Here we propose DeepSEM, a deep generative model that can jointly infer GRNs and biologically meaningful representation of single-cell RNA sequencing (scRNA-seq) data. In particular, we developed a neural network version of the structural equation model (SEM) to explicitly model the regulatory relationships among genes. Benchmark results show that DeepSEM achieves comparable or better performance on a variety of single-cell computational tasks, such as GRN inference, scRNA-seq data visualization, clustering and simulation, compared with the state-of-the-art methods. In addition, the gene regulations predicted by DeepSEM on cell-type marker genes in the mouse cortex can be validated by epigenetic data, which further demonstrates the accuracy and efficiency of our method. DeepSEM can provide a useful and powerful tool to analyze scRNA-seq data and infer a GRN.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
We provide all datasets generated or analyzed during this study. The gene experimental scRNA-seq datasets were downloaded from Gene Expression Omnibus with the accession numbers GSE81252 (hHEP dataset65), GSE75748 (hESC dataset66), GSE98664 (mESC dataset62), GSE48968 (mDC dataset63), GSE81682 (mHSC dataset64), GSE115746 (mouse cortex dataset33), GSE60361 (Zeisel dataset41), GSE85241 (Muraro dataset78), GSE81861 (Li dataset79), and GSE45719 (Deng dataset80). The other experimental scRNA-seq dataset were downloaded from ArrayExpress with the accession number E-MTAB-5061 (Segerstolpe dataset81), NCBI Sequence Read Archive (SRA) with accession number SRP041736 (Pollen dataset42), GitHub repositories (https://github.com/LuyiTian/sc_mixology) (CellBench dataset82) and the website for x10genomics (https://support.10xgenomics.com/single-cell-gene-expression/datasets/) (PBMC dataset43). The scATAC-seq and snmC-seq for mouse cortex were downloaded from Gene Expression Omnibus with the accession numbers GSE126724 (scATAC-seq35) and GSE97179 (snmC-seq34). More information for these datasets could be found in Methods. We also summarize the accession and download links in Supplementary Tables 1, 2, 5 and 9. Source Data for Figs. 3–6 are available with this manuscript.
Code availability
The codes generated during this study are available on GitHub (https://github.com/HantaoShu/DeepSEM) and in Zenodo83.
References
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Hashimshony, T. et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-seq. Genome Biol. 17, 77 (2016).
Wagner, A., Regev, A. & Yosef, N. Revealing the vectors of cellular identity with single-cell genomics. Nat. Biotechnol. 34, 1145–1160 (2016).
Kharchenko, P. V., Silberstein, L. & Scadden, D. T. Bayesian approach to single-cell differential expression analysis. Nat. Methods 11, 740–742 (2014).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Eraslan, G., Simon, L. M., Mircea, M., Mueller, N. S. & Theis, F. J. Single-cell RNA-seq denoising using a deep count autoencoder. Nat. Commun. 10, 390 (2019).
Cuomo, A. S. E. et al. Single-cell RNA-sequencing of differentiating iPS cells reveals dynamic genetic effects on gene expression. Nat. Commun. 11, 810 (2020).
Olsson, A. et al. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537, 698–702 (2016).
Sharma, A. et al. Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell 183, 377–394.e21 (2020).
Arisdakessian, C., Poirion, O., Yunits, B., Zhu, X. & Garmire, L. X. DeepImpute: an accurate, fast, and scalable deep neural network method to impute single-cell RNA-seq data. Genome Biol. 20, 211 (2019).
Wang, T. et al. BERMUDA: a novel deep transfer learning method for single-cell RNA sequencing batch correction reveals hidden high-resolution cellular subtypes. Genome Biol. 20, 165 (2019).
Li, X. et al. Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis. Nat. Commun. 11, 2338 (2020).
Huynh-Thu, V. A., Irrthum, A., Wehenkel, L. & Geurts, P. Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 5, e12776 (2010).
Chan, T. E., Stumpf, M. P. H. & Babtie, A. C. Gene regulatory network inference from single-cell data using multivariate information measures. Cell Syst. 5, 251–267.e3 (2017).
Matsumoto, H. et al. SCODE: an efficient regulatory network inference algorithm from single-cell RNA-seq during differentiation. Bioinformatics 33, 2314–2321 (2017).
Papili Gao, N., Ud-Dean, S. M. M., Gandrillon, O. & Gunawan, R. SINCERITIES: inferring gene regulatory networks from time-stamped single cell transcriptional expression profiles. Bioinformatics 34, 258–266 (2018).
Moerman, T. et al. GRNBoost2 and Arboreto: efficient and scalable inference of gene regulatory networks. Bioinformatics 35, 2159–2161 (2019).
Kamimoto, K., Hoffmann, C. M. & Morris, S. A. CellOracle: dissecting cell identity via network inference and in silico gene perturbation. Preprint at bioRxiv https://doi.org/10.1101/2020.02.17.947416 (2020).
Kim, S. ppcor: an R package for a fast calculation to semi-partial correlation coefficients. Commun. Stat. Appl. Methods 22, 665–674 (2015).
Yu, Y., Jie, C., Tian, G. & Mo, Y. DAG-GNN: DAG structure learning with graph neural networks. In Proceedings of the 36th International Conference on Machine Learning 7154–7163 (ICML, 2019).
Lin, C., Jain, S., Kim, H. & Bar-Joseph, Z. Using neural networks for reducing the dimensions of single-cell RNA-seq data. Nucleic Acids Res. 45, e156 (2017).
Higgins, I. et al. beta-VAE: learning basic visual concepts with a constrained variational framework. In Proceedings of the 5th International Conference on Learning Representations (ICML, 2017).
Zhao, A., Balakrishnan, G., Durand, F., Guttag, J. V. & Dalca, A. V. Data augmentation using learned transformations for one-shot medical image segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 8543–8553 (IEEE, 2019).
Lotfollahi, M., Wolf, F. A. & Theis, F. J. scGen predicts single-cell perturbation responses. Nat. Methods 16, 715–721 (2019).
Wang, X., Ghasedi Dizaji, K. & Huang, H. Conditional generative adversarial network for gene expression inference. Bioinformatics 34, i603–i611 (2018).
Marouf, M. et al. Realistic in silico generation and augmentation of single-cell RNA-seq data using generative adversarial networks. Nat. Commun. 11, 166 (2020).
Pratapa, A., Jalihal, A. P., Law, J. N., Bharadwaj, A. & Murali, T. M. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods 17, 147–154 (2020).
Moore, L. D., Le, T. & Fan, G. DNA methylation and its basic function. Neuropsychopharmacology (2013).
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Keilwagen, J., Posch, S. & Grau, J. Accurate prediction of cell type-specific transcription factor binding. Genome Biol. 20, 9 (2019).
Funk, C. C. et al. Atlas of transcription factor binding sites from ENCODE DNase hypersensitivity data across 27 tissue types. Cell Rep. 32, 108029 (2020).
Tasic, B. et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018).
Luo, C. et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science 357, 600–604 (2017).
Fang, R. et al. Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. 12, 1337 (2021).
Dong, J. et al. Enhancing single-cell cellular state inference by incorporating molecular network features. Preprint at bioRxiv (2019).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Li, X. et al. Network embedding-based representation learning for single cell RNA-seq data. Nucleic Acids Res. 45, e166–e166 (2017).
Cahan, P. et al. CellNet: network biology applied to stem cell engineering. Cell 158, 903–915 (2014).
Morris, S. A. et al. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell 158, 889–902 (2014).
Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015).
Pollen, A. A. et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 32, 1053–1058 (2014).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Pierson, E. & Yau, C. ZIFA: dimensionality reduction for zero-inflated single-cell gene expression analysis. Genome Biol. 16, 241 (2015).
Jolliffe, I. T. in Principal Component Analysis (ed. Jolliffe, I. T.) 115–128 (Springer, 1986).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. https://doi.org/10.1038/nbt.4314 (2019).
Heiser, C. N. & Lau, K. S. A quantitative framework for evaluating single-cell data structure preservation by dimensionality reduction techniques. Cell Rep. 31, 107576 (2020).
Viñas, R., Andrés-Terré, H., Liò, P. & Bryson, K. Adversarial generation of gene expression data. Bioinformatics https://doi.org/10.1093/bioinformatics/btab035 (2021).
Bollen, K. A. Structural Equations with Latent Variables (John Wiley & Sons, 1989).
Haavelmo, T. The statistical implications of a system of simultaneous equations. Econometrica 11, 1–12 (1943).
King, M., Goldberger, A. S. & Duncan, O. D. Structural equation models in the social sciences. Econ. J. 84, 212–214 (1974).
Duarte, C. W., Klimentidis, Y. C., Harris, J. J., Cardel, M. & Fernández, J. R. A hybrid Bayesian network/structural equation (BN/SEM) modeling approach for detecting physiological networks for obesity-related genetic variants. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine 696–702 (IEEE, 2012).
Yoo, C. & Oh, S. Combining structure equation model with Bayesian networks for predicting with high accuracy of recommending surgery for better survival in Benign prostatic hyperplasia patients. In 20th International Congress on Modelling and Simulation-Adapting to Change 2029–2033 (Modelling and Simulation Society of Australia and New Zealand, 2013).
Zheng, X., Aragam, B., Ravikumar, P. & Xing, E. P. DAGs with NO TEARS: continuous optimization for structure learning. In Proceedings of the 32nd International Conference on Neural Information Processing Systems 9492–9503 (IEEE, 2018).
Luo, Y., Peng, J. & Ma, J. When causal inference meets deep learning. Nat. Mach. Intell. 2, 426–427 (2020).
Kingma, D. P. & Welling, M. An introduction to variational autoencoders. Found Trends Mach. Learn. 12, 307–392 (2019).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. In Proceedings of the 3th International Conference on Learning Representations (ICLR, 2015).
Friedman, J., Hastie, T. & Tibshirani, R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics 9, 432–441 (2008).
Tieleman, T. & Hinton, G. Lecture 6.5-rmsprop, Coursera: Neural Networks for Machine Learning Technical Report (Univ. Toronto, 2012).
He, K., Zhang, X., Ren, S. & Sun, J. Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. In Proceedings of the IEEE International Conference on Computer Vision 1026–1034 (IEEE, 2015).
Hayashi, T. et al. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat. Commun. 9, 619 (2018).
Shalek, A. K. et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510, 363–369 (2014).
Nestorowa, S. et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128, e20–e31 (2016).
Camp, J. G. et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538 (2017).
Chu, L.-F. et al. Single-cell RNA-seq reveals novel regulators of human embryonic stem cell differentiation to definitive endoderm. Genome Biol. 17, 173 (2016).
ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Davis, C. A. et al. The encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 46, D794–D801 (2018).
Oki, S. et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 19, e46255 (2018).
Xu, H. et al. ESCAPE: database for integrating high-content published data collected from human and mouse embryonic stem cells. Database 2013, bat045 (2013).
Garcia-Alonso, L., Holland, C. H., Ibrahim, M. M., Turei, D. & Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 29, 1363–1375 (2019).
Liu, Z.-P., Wu, C., Miao, H. & Wu, H. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database 2015, bav095 (2015).
Han, H. et al. TRRUST: a reference database of human transcriptional regulatory interactions. Sci. Rep. 5, 11432 (2015).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Fornes, O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020).
Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011).
Muraro, M. J. et al. A Single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394.e3 (2016).
Li, H. et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 49, 708–718 (2017).
Deng, Q., Ramsköld, D., Reinius, B. & Sandberg, R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343, 193–196 (2014).
Segerstolpe, Å. et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607 (2016).
Tian, L. et al. Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments. Nat. Methods 16, 479–487 (2019).
Shu, H. et al. Code for paper ‘Modeling gene regulatory networks using neural network architectures’. Zenodo https://doi.org/10.5281/zenodo.4915754 (2021).
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (61872216, 81630103), the Turing AI Institute of Nanjing to J. Zeng. We also acknowledge the National Natural Science Foundation of China (31900862) for funding support to D.Z..
Author information
Authors and Affiliations
Contributions
J. Zeng and J.M. designed the study and developed the conceptual ideas. H.S. and Q.L. implemented the main algorithms. H.S. performed the model training and experimental validation task. H.S. and J. Zhou collected all the input data sources and interpreted the results. H.S., J. Zhou, H.L., D.Z., J. Zeng and J.M. wrote the manuscript with support from all authors.
Corresponding authors
Ethics declarations
Competing interests
J. Zeng is founder and CTO of Silexon AI Technology Co. Ltd. and has an equity interest. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Computational Science thanks Jun Ding, Yafei Lyu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Handling editor: Fernando Chirigati, in collaboration with the Nature Computational Science team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–17, Sections 1–7 and Tables 1–10.
Source data
Source Data Fig. 3
The corresponding raw data file for Fig. 3.
Source Data Fig. 4
The corresponding raw data file for Fig. 4.
Source Data Fig. 5
The corresponding raw data file for Fig. 5.
Source Data Fig. 6
The corresponding raw data file for Fig. 6.
Rights and permissions
About this article
Cite this article
Shu, H., Zhou, J., Lian, Q. et al. Modeling gene regulatory networks using neural network architectures. Nat Comput Sci 1, 491–501 (2021). https://doi.org/10.1038/s43588-021-00099-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-021-00099-8
This article is cited by
-
A novel interpretable deep transfer learning combining diverse learnable parameters for improved T2D prediction based on single-cell gene regulatory networks
Scientific Reports (2024)
-
Inference of Gene Regulatory Networks Based on Multi-view Hierarchical Hypergraphs
Interdisciplinary Sciences: Computational Life Sciences (2024)
-
EnsInfer: a simple ensemble approach to network inference outperforms any single method
BMC Bioinformatics (2023)
-
A systematic review of biologically-informed deep learning models for cancer: fundamental trends for encoding and interpreting oncology data
BMC Bioinformatics (2023)
-
Diffusion models in bioinformatics and computational biology
Nature Reviews Bioengineering (2023)