Abstract
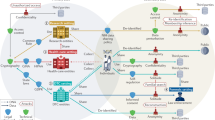
The growing number of health-data breaches, the use of genomic databases for law enforcement purposes and the lack of transparency of personal genomics companies are raising unprecedented privacy concerns. To enable a secure exploration of genomic datasets with controlled and transparent data access, we propose a citizen-centric approach that combines cryptographic privacy-preserving technologies, such as homomorphic encryption and secure multi-party computation, with the auditability of blockchains. Our open-source implementation supports queries on the encrypted genomic data of hundreds of thousands of individuals, with minimal overhead. We show that real-world adoption of our system alleviates widespread privacy concerns and encourages data access sharing with researchers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
To evaluate the performance of our system, we used open-access genomic and clinical data from The Cancer Genome Atlas (TCGA). The initial dataset contained one million genetic variants distributed across 8,000 individuals that were obtained by downloading and merging genomic data for the 32 studies of the TCGA PanCancer Atlas from https://www.cbioportal.org/, and are made available at https://github.com/ldsec/projects-data/tree/master/medco/datasets/genomic/tcga_cbio. We synthetically augmented this initial dataset to obtain the final test dataset of 28 billion genetic variants distributed across 150,000 individuals (each individual has a number of genetic variants, ranging from 15,000 to 200,000). This dataset is very large (in the order of tens of terabytes), so we cannot host it online. Instructions for reproducing the dataset augmentation are provided in the GitHub repository. It also includes a small demo dataset (https://github.com/ldsec/ccgd-platform) that is sufficient to test the code. If access to the large dataset that was used for the performance benchmarks is required, contact juan.troncoso-pastoriza@epfl.ch.
Code availability
The code is available on GitHub at https://github.com/ldsec/ccgd-platform. The code is archived on Zenodo at https://doi.org/10.5281/zenodo.455116513.
References
Regalado, A. More than 26 million people have taken an at-home ancestry test. MIT Technology Review (11 February 2019).
Farr, C. 23andMe lays off 100 people as DNA test sales decline, CEO says she was ‘surprised’ to see market turn. CNBC https://www.cnbc.com/2020/01/23/23andme-lays-off-100-people-ceo-anne-wojcicki-explains-why.html (2020).
Farr, C. Ancestry to lay off 6% of workforce because of a slowdown in the consumer DNA-testing market. CNBC https://www.cnbc.com/2020/02/05/ancestry-layoffs-of-6percent-100-people-amid-dna-test-slowdown.html (2020).
Huang, Z., Ayday, E., Fellay, J., Hubaux, J. & Juels, A. GenoGuard: protecting genomic data against brute-force attacks. In Proc. 2015 IEEE Symposium on Security and Privacy 447–462 (IEEE, 2015).
Cho, H., Wu, D. J. & Berger, B. Secure genome-wide association analysis using multiparty computation. Nat. Biotechnol. 36, 547–551 (2018).
Jagadeesh, K. A., Wu, D. J., Birgmeier, J. A., Boneh, D. & Bejerano, G. Deriving genomic diagnoses without revealing patient genomes. Science 357, 692–695 (2017).
Bogdanov, D., Laur, S. & Willemson, J. Sharemind: a framework for fast privacy-preserving computations. In Proc. Computer Security ESORICS 2008 192–206 (Springer, 2008).
Boura, C. et al. in Financial Cryptography and Data Security 183–202 (Springer, 2018).
Raisaro, J. L. et al. MedCo: enabling secure and privacy-preserving exploration of distributed clinical and genomic data. IEEE/ACM Trans. Comput. Biol. Bioinform. 16, 1328–1341 (2018).
Grishin, D., Obbad, K. & Church, G. M. Data privacy in the age of personal genomics. Nat. Biotechnol. 37, 1115–1117 (2019).
Froelicher, D. et al. UnLynx: a decentralized system for privacy-conscious data sharing. Proc. Priv. Enhancing Technol. 2017, 232–250 (2017).
Luterbacher, C. EPFL software to enable secure data-sharing for hospitals. EPFL (2 April 2002).
de Sa Sousa, J. A. G., Misbach, M., Quinn, K., Pastoriza, J. R. T. & Grishin, D. ldsec/ccgd-platform: Citizen-Centered Genomic Discovery Platform (Zenodo, 2021); https://doi.org/10.5281/zenodo.4551165
Acknowledgements
We are grateful to the Bitfury Exonum engineering team for running part of the performance measurements described in this paper. We also thank C. Redin from Lausanne University Hospital, J. Aach from Harvard Medical School and A. Pyrgelis from EPFL for their useful feedback and constructive comments on the manuscript. This work was supported in part by grant no. 2017-201 (DPPH) of the Swiss strategic focus area Personalized Health and Related Technologies (PHRT) and by grant no. 2018-522 (MedCo) of the PHRT and the Swiss Personalized Health Network (SPHN). The work was also supported by Nebula Genomics.
Author information
Authors and Affiliations
Contributions
D.G., J.L.R. and J.T.P. conceived the study. D.G., J.L.R., J.T.P. and J.P.H. wrote the manuscript. D.G., J.L.R., J.T.P., K.O., J.G., M.M., K.Q. and J.S. implemented the algorithms described in the Methods and performed the benchmark experiments. All authors discussed and reviewed the manuscript at all stages. J.F., G.M.C. and J.P.H. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
D.G., K.O., K.Q. and G.M.C. are affiliated with Nebula Genomics. J.L.R., J.T.P., M.M., J.S., J.F. and J.P.H. declare no competing interests.
Additional information
Peer review information Nature Computational Science thanks Fida Dankar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Ananya Rastogi was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Security and privacy background, example of secure distributed aggregate-data analysis, Supplementary Results (Figs. 1–9) and Supplementary Methods (Figs. 10–18).
Source data
Source Data Fig. 1
Survey responses.
Source Data Fig. 3
Code benchmarks.
Rights and permissions
About this article
Cite this article
Grishin, D., Raisaro, J.L., Troncoso-Pastoriza, J.R. et al. Citizen-centered, auditable and privacy-preserving population genomics. Nat Comput Sci 1, 192–198 (2021). https://doi.org/10.1038/s43588-021-00044-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-021-00044-9
This article is cited by
-
Data breaches in healthcare: security mechanisms for attack mitigation
Cluster Computing (2024)
-
Sociotechnical safeguards for genomic data privacy
Nature Reviews Genetics (2022)
-
Data privacy through participant empowerment
Nature Computational Science (2021)