Abstract
The aging brain exhibits a region-specific reduction in synapse number and plasticity. Although astrocytes play central roles in regulating synapses, it is unclear how changes in astrocytes contribute to age-dependent cognitive decline and vulnerability to neurodegenerative diseases. Here, we identified a unique astrocyte subtype that exhibits dysregulated autophagy and morphology in aging hippocampus. In these autophagy-dysregulated astrocytes (APDAs), autophagosomes abnormally accumulate in swollen processes, impairing protein trafficking and secretion. We found that reduced mammalian target of rapamycin (mTOR) and proteasome activities with lysosomal dysfunction generate APDAs in an age-dependent manner. Secretion of synaptogenic molecules and astrocytic synapse elimination were significantly impaired in APDAs, suggesting that APDAs have lost their ability to control synapse number and homeostasis. Indeed, excitatory synapses and dendritic spines associated with APDAs were significantly reduced. Finally, we found that mouse brains with Alzheimer’s disease showed a significantly accelerated increase in APDAs, suggesting potential roles for APDAs in age- and Alzheimer’s disease-related cognitive decline and synaptic pathology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The raw bulk RNA-seq data are available at the Gene Expression Omnibus under the accession code GSE183042. Source data is provided with this document. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
Chung, W. S., Allen, N. J. & Eroglu, C. Astrocytes control synapse formation, function, and elimination. Cold Spring Harb. Perspect. Biol. 7, a020370 (2015).
Clarke, L. E. & Barres, B. A. Emerging roles of astrocytes in neural circuit development. Nat. Rev. Neurosci. 14, 311–321 (2013).
Christopherson, K. S. et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 120, 421–433 (2005).
Kucukdereli, H. et al. Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC. Proc. Natl Acad. Sci. USA 108, E440–E449 (2011).
Singh, S. K. et al. Astrocytes assemble thalamocortical synapses by bridging NRX1alpha and NL1 via Hevin. Cell 164, 183–196 (2016).
Allen, N. J. et al. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature 486, 410–414 (2012).
Chung, W. S. et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504, 394–400 (2013).
Yang, J. et al. Astrocytes contribute to synapse elimination via type 2 inositol 1,4,5-trisphosphate receptor-dependent release of ATP. eLife 5, e15043 (2016).
Matias, I., Morgado, J. & Gomes, F. C. A. Astrocyte heterogeneity: impact to brain aging and disease. Front. Aging Neurosci. 11, 59 (2019).
Boisvert, M. M., Erikson, G. A., Shokhirev, M. N. & Allen, N. J. The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 22, 269–285 (2018).
Clarke, L. E. et al. Normal aging induces A1-like astrocyte reactivity. Proc. Natl Acad. Sci. USA 115, E1896–E1905 (2018).
Morrison, J. H. & Baxter, M. G. The ageing cortical synapse: hallmarks and implications for cognitive decline. Nat. Rev. Neurosci. 13, 240–250 (2012).
Scheff, S. W., Price, D. A., Schmitt, F. A. & Mufson, E. J. Hippocampal synaptic loss in early Alzheimer’s disease and mild cognitive impairment. Neurobiol. Aging 27, 1372–1384 (2006).
Rodriguez, J. J. et al. Complex and region-specific changes in astroglial markers in the aging brain. Neurobiol. Aging 35, 15–23 (2014).
Zamanian, J. L. et al. Genomic analysis of reactive astrogliosis. J. Neurosci. 32, 6391–6410 (2012).
Liddelow, S. A. et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017).
Batiuk, M. Y. et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 11, 1220 (2020).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Morimoto, R. I. & Cuervo, A. M. Proteostasis and the aging proteome in health and disease. J. Gerontol. A Biol. Sci. Med. Sci. 69, S33–S38 (2014).
Vilchez, D., Saez, I. & Dillin, A. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat. Commun. 5, 5659 (2014).
Kwon, Y. T. & Ciechanover, A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 42, 873–886 (2017).
Lee, J. H. et al. Astrocytes phagocytose adult hippocampal synapses for circuit homeostasis. Nature 590, 612–617 (2021).
Basak, J. M., Verghese, P. B., Yoon, H., Kim, J. & Holtzman, D. M. Low-density lipoprotein receptor represents an apolipoprotein E-independent pathway of Aβ uptake and degradation by astrocytes. J. Biol. Chem. 287, 13959–13971 (2012).
O’Kane, R. L., Martinez-Lopez, I., DeJoseph, M. R., Vina, J. R. & Hawkins, R. A. Na(+)-dependent glutamate transporters (EAAT1, EAAT2, and EAAT3) of the blood-brain barrier. A mechanism for glutamate removal. J. Biol. Chem. 274, 31891–31895 (1999).
Kocaturk, N. M. & Gozuacik, D. Crosstalk between mammalian autophagy and the ubiquitin-proteasome system. Front. Cell Dev. Biol. 6, 128 (2018).
Johnson, S. C., Rabinovitch, P. S. & Kaeberlein, M. mTOR is a key modulator of ageing and age-related disease. Nature 493, 338–345 (2013).
Garza-Lombo, C. & Gonsebatt, M. E. Mammalian target of rapamycin: its role in early neural development and in adult and aged brain function. Front. Cell Neurosci. 10, 157 (2016).
Dikic, I. & Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018).
Castillo, K. et al. Measurement of autophagy flux in the nervous system in vivo. Cell Death Dis. 4, e917 (2013).
Leeman, D. S. et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283 (2018).
Ge, D. et al. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy 10, 957–971 (2014).
Wei, L. et al. A butyrolactone derivative 3BDO alleviates memory deficits and reduces amyloid-beta deposition in an AbetaPP/PS1 transgenic mouse model. J. Alzheimers Dis. 30, 531–543 (2012).
Inoue, M. et al. SCRG1 suppresses LPS-induced CCL22 production through ERK1/2 activation in mouse macrophage Raw264.7 cells. Mol. Med. Rep. 15, 4069–4076 (2017).
Mostany, R. et al. Altered synaptic dynamics during normal brain aging. J. Neurosci. 33, 4094–4104 (2013).
Habib, N. et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23, 701–706 (2020).
Jankowsky, J. L. et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum. Mol. Genet. 13, 159–170 (2004).
Misrani, A., Tabassum, S. & Yang, L. Mitochondrial dysfunction and oxidative stress in Alzheimer’s disease. Front. Aging Neurosci. 13, 617588 (2021).
Sakai, K., Fukuda, T. & Iwadate, K. Beading of the astrocytic processes (clasmatodendrosis) following head trauma is associated with protein degradation pathways. Brain Inj. 27, 1692–1697 (2013).
Early, A. N., Gorman, A. A., Van Eldik, L. J., Bachstetter, A. D. & Morganti, J. M. Effects of advanced age upon astrocyte-specific responses to acute traumatic brain injury in mice. J. Neuroinflammation 17, 115 (2020).
Hulse, R. E., Winterfield, J., Kunkler, P. E. & Kraig, R. P. Astrocytic clasmatodendrosis in hippocampal organ culture. Glia 33, 169–179 (2001).
Ryu, H. J., Kim, J. E., Yeo, S. I. & Kang, T. C. p65/RelA-Ser529 NF-kappaB subunit phosphorylation induces autophagic astroglial death (Clasmatodendrosis) following status epilepticus. Cell Mol. Neurobiol. 31, 1071–1078 (2011).
Vainchtein, I. D. et al. Astrocyte-derived interleukin-33 promotes microglial synapse engulfment and neural circuit development. Science 359, 1269–1273 (2018).
Sanmarco, L. M. et al. Gut-licensed IFNgamma(+) NK cells drive LAMP1(+)TRAIL(+) anti-inflammatory astrocytes. Nature 590, 473–479 (2021).
Ito, M. et al. Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature 565, 246–250 (2019).
Brewer, G. J. & Torricelli, J. R. Isolation and culture of adult neurons and neurospheres. Nat. Protoc. 2, 1490–1498 (2007).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Lois, C., Hong, E. J., Pease, S., Brown, E. J. & Baltimore, D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295, 868–872 (2002).
Guo, P. et al. A simplified purification method for AAV variant by polyethylene glycol aqueous two-phase partitioning. Bioengineered 4, 103–106 (2013).
Foo, L. C. et al. Development of a method for the purification and culture of rodent astrocytes. Neuron 71, 799–811 (2011).
Byun, Y. C. & Chung, W. S. A novel in vitro live-imaging assay of astrocyte-mediated phagocytosis using pH indicator-conjugated synaptosomes. J. Vis. Exp. 132, 56647 (2018).
Gilles, J. F., Dos Santos, M., Boudier, T., Bolte, S. & Heck, N. DiAna, an ImageJ tool for object-based 3D co-localization and distance analysis. Methods 115, 55–64 (2017).
Binley, K. E., Ng, W. S., Tribble, J. R., Song, B. & Morgan, J. E. Sholl analysis: a quantitative comparison of semi-automated methods. J. Neurosci. Methods 225, 65–70 (2014).
Risher, W. C., Ustunkaya, T., Singh Alvarado, J. & Eroglu, C. Rapid Golgi analysis method for efficient and unbiased classification of dendritic spines. PLoS One 9, e107591 (2014).
Howard, V. & Reed, M. G. Unbiased Stereology: Three-dimensional Measurement in Microscopy (BIOS Scientific, 1998).
Park, J. et al. Microglial MERTK eliminates phosphatidylserine-displaying inhibitory post-synapses. EMBO J. 40, e107121 (2021).
Acknowledgements
We thank all members in Chung’s laboratory for helpful discussion. This work was supported by grants from the Samsung Science and Technology Foundation (SSTF-BA1701-18, W-.S.C.). This work was also supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (Ministry of Science and ICT, MSIT) [2022R1A2C1009376 (J.Y.M.)], KBRI basic research program [22-BR-01-03 (J.Y.M.)] funded by MSIT, the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) and Korea Dementia Research Center (KDRC), funded by the Ministry of Health and Welfare (MOHW) and MSIT, Republic of Korea (HU20C0290) and International Collaboration Initiative grant (KAIST-N11210255) supported by KAIST. scRNA-seq experiments were conducted with help from Macrogen and Amyloid Solution Inc. Imaris software was provided by KAIST Bio Core Center.
Author information
Authors and Affiliations
Contributions
W-.S.C., E.L. and Y-.J.J. designed the project. E.L performed all the experiments including primary cell isolation, plasmid cloning, virus production, cell culture, immunohistochemistry, confocal imaging and western blot. Y.-J.C. performed stereotaxic injection with virus and drugs. S.Y.L. provided the brain slices injected with virus and performed behavior analysis. Y.R.P. and J.Y.M. performed electron microscopy experiments. S.L. and C.H.K. performed scRNA-seq analysis. W-.S.C. and E.L. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Andras Lakatos, Cagla Eroglu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
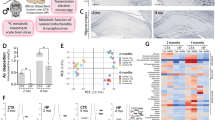
Extended Data Fig. 1 Investigation of vacuolated astrocytes in the young and aged hippocampus.
a, Schematic illustration of the scRNA-seq experiments. b, Comparison of upregulated genes in astrocyte clusters from 2-year-old hippocampi with scRNA-seq dataset obtained from 2-month-old hippocampus. Note that characteristic gene expression changes in cluster 3 and 6 are unique to the aged hippocampus. c, A UMAP plot of single cells from the 2-month-old hippocampi (left panel). Bar graph showing the relative percentages of five different astrocyte clusters (right panel). d, Violin plots showing the expression of Scrg1, Gstm1 and Cldn10 in cluster 2, 3 and 6 from 2-year-old hippocampi. e, Quantification of SCRG1, GSTM1 and CLDN10 areas normalized by astrocyte area in 4-, 6-, 9- and 18-month-old WT hippocampi (n = 3, dots on the bar graph indicate data obtained from individual ROIs). f,g,i Representative confocal z-stack images of normal and vacuolated astrocytes obtained from 18-month-old WT hippocampi stained for s100β (green), Cx43 (f, red), APOE (g, red) and HIF1α (i, red). These proteins are accumulated in the swollen processes of vacuolated astrocytes. Scale bar, 10 μm. h,j–m, Quantification of vimentin (h), MEGF10 (j), Cx43 (k), APOE (l) and HIF1α (m) areas normalized by astrocyte area in 18-month-old WT hippocampi (n = 5 mice, dots on the bar graphs represent data obtained from individual cells). e, One-way ANOVA followed by Tukey’s multiple comparison test. The P-values are indicated in the source data file. h,j–m, Two-tailed Student’s t-test. P = 0.21 (h), **P = 0.001 (j), ****P < 0.0001 (k), *P = 0.015 (l), ****P < 0.0001 (m). ns, not significant. Mean ± s.e.m.
Extended Data Fig. 2 Selective appearance of vacuolated astrocytes in the hippocampus.
a, Representative confocal z-stack images of 18-month-old hippocampi stained for nuclei (DAPI, blue), s100β (green) and SCRG1 (red). Astrocytes with upregulated SCRG1 expression were found only in the SR and SLM layers of the hippocampus. Scale bar, 50 μm. b, Quantification of the number of vacuolated astrocytes normalized to the total number of astrocytes in the imaging fields (n = 3, dots on the bar graph indicate data obtained from individual ROIs). c, Representative magnified confocal z-stack images showing the absence of SCRG1 (red)-expressing cells in other brain regions, such as the cortex, thalamus and hypothalamus. d, Representative magnified confocal z-stack images showing the absence of SCRG1 (red) expression in other cell types, such as microglia (Iba1, upper), neurons (NeuN, middle) and oligodendrocyte precursor cells (PDGFRα, lower). For both c and d, the left panels show representative images taken at ×40 magnification (scale bar, 50 μm), and the right panels show representative single astrocytes (s100β, green) taken at ×120 magnification (scale bar, 10 μm). DAPI (blue) stains the nucleus. e, Sholl analysis of normal (left) and vacuolated (right) astrocytic processes stained for s100β (green). The concentric circles were placed at 1μm apart, with the astrocytic soma in the middle (white lines). f, Quantification of the number of nodes where the processes intersect with the concentric circles (n = 3, ROIs obtained from twelve images). b, One-way ANOVA followed by Tukey’s multiple comparisons test. f, Two-way ANOVA followed by Sidak’s multiple comparisons test. The P-values are indicated in the source data file (b,f). Mean ± s.e.m.
Extended Data Fig. 3 Vacuolated astrocytes were neither apoptotic nor necrotic.
a, Representative confocal z-stack images of 18-month-old hippocampi showing normal astrocytes (upper panels) and vacuolated astrocytes (middle panels). Autophagosomes (p62, cyan) accumulated in vacuolated astrocytes (s100β, green). However, these vacuolated astrocytes were not positive for activated caspase 3 signals (red). Lower panels were representative confocal images from 4-month-old cortex injected with ATP stained for nuclei (DAPI, blue), GFAP (green) and active caspase-3 (red). Scale bar, 10 μm. b, TUNEL staining of brain sections from 18-month-old WT mice along with a positive control of DNase I-treated brain sections (lower panels) stained for s100β (green) and p62 (cyan). APDA marked with a white arrow was not positive for TUNEL staining (red). Scale bar, 50 μm. c, Representative confocal z-stack images of 18-month-old hippocampi (upper panels) stained for p62 (cyan), s100β (green) and HMGB1 (red). APDA marked with a white arrow was not positive for HMGB1, a marker for necrosis. Lower panels were representative confocal images from 4-month-old cortex injected with ATP stained for nuclei (DAPI, blue), s100β (green) and HMGB1 (red). Scale bar, 50 μm. Dotted lines indicate APDA’s territory (b–c).
Extended Data Fig. 4 Age dependent appearance of APDAs by mTOR and proteasome inhibition.
a–d, Quantification of MEGF10 (a,c) and HIF1α (b,d) areas normalized by astrocyte area in the imaging fields. Normal astrocytes and APDAs were obtained from 9-month-old Aldh1l1-eGFP hippocampi injected with rapamycin (Rapa, a,b), and MG132 and CQ (MG + CQ, c,d). MEGF10 (a,c) and HIF1α (b,d) were highly upregulated in APDAs compared to normal astrocytes. e, Quantification showing the relative number of APDAs per total astrocytes in the SR hippocampal layers of 9-month-old Aldh1l1-eGFP hippocampi injected with CQ (n = 3, dots on the bar graphs indicate data obtained from individual ROIs). (n = 3 mice, dots on the bar graphs indicate data obtained from individual ROIs). f, Representative confocal z-stack images of 4-month-old Aldh1l1-eGFP hippocampi injected with vehicle (Veh, left), rapamycin (Rapa, middle), rapamycin and chloroquine (Rapa+CQ, right) stained for nuclei (DAPI, blue), GFP (green) and SCRG1 (red). Scale bar, 50 μm. g, Quantification of the relative number of APDAs per the total number of astrocytes in the SR hippocampal layers (n = 4 mice; dots on the bar graphs indicate data obtained from individual ROIs). h, Representative confocal z-stack images of 4-month-old Aldh1l1-eGFP hippocampi injected with vehicle (Veh, left), MG132 (MG, middle), MG132 plus chloroquine (MG + CQ, right) and stained for nuclei (DAPI, blue), GFP (green) and SCRG1 (red). Scale bar, 50 μm. White arrows indicate APDAs in the hippocampus injected with MG + CQ. i, Quantification showing the relative number of APDAs per the total number of astrocytes in the SR hippocampal layers (n = 3 mice; dots on the bar graphs indicate data obtained from individual ROIs). j, Representative magnified confocal z-stack images showing the absence of SCRG1 (red)-expressing cells in other brain regions, such as the cortex, thalamus and hypothalamus of 9-month-old Aldh1l1-eGFP hippocampi injected with MG132 and CQ. k–l Quantification of Cx43 (k) and MEGF10 (l) areas normalized by astrocyte area. Cx43 and MEGF10 were highly upregulated in 9-month-old Aldh1l1-eGFP mice injected with MG132 and CQ compared to 4-month-old mice (n = 3 for 9-month-old and n = 4 for 4-month-old mice, dots on the bar graphs indicate data obtained from individual ROIs). a–e,k,l two-tailed Student’s t-test. ***P = 0.0009 (a), ****P < 0.0001 (b), P = 0.1234 (c), ****P < 0.0001 (d), ****P < 0.0001 (e), ****P < 0.0001 (k), *P < 0.023 (l). g,i One-way ANOVA followed by Tukey’s multiple comparisons test. The P-values are indicated in the source data file (g,i). Mean ± s.e.m.
Extended Data Fig. 5 In vivo secretion of SCRG1, Hevin and SPARC from normal astrocytes and APDAs.
a, Schematic diagram of a lentiviral construct expressing SCRG1-HA under the Gfap promoter injected into the hippocampal CA1 region. b, Representative confocal z-stack images of contralateral and ipsilateral hippocampal regions injected with virus expressing SCRG1-HA. Extracellular SCRG1-HA (HA, green) signal was found outside of SCRG1-HA-expressing astrocytes (GFAP, red) on the ipsilateral side of the hippocampus. Scale bar, 50 μm. c, Enlarged images of the white boxes shown in b. The signal of SCRG1-HA (HA, green) can be found in regions (i) where there were no SCRG1-HA-expressing astrocytes (ii, images of astrocytes expressing SCRG1-HA). Scale bar, 50 μm. d,f Representative confocal z-stack images of normal astrocytes and APDAs (s100β, green) stained for Hevin (d, red) and SPARC (f, red) in the 18-month-old hippocampi. e,g, The number of extracellular Hevin (e) and SPARC (g) puncta were plotted based on the distance from the soma. (n = 12 cells examined over three mice) Unpaired Student’s t-test. ****P < 0.0001 (e,g), Mean ± s.e.m.
Extended Data Fig. 6 The number of inhibitory synapses in 18-month-old mice and the number of excitatory synapses in 9-month-old mice injected with rapamycin and CQ are not changed near APDAs.
a, Representative confocal z-stack images of inhibitory synapses stained for VGAT (red) and Gephyrin (green) around normal astrocytes and APDAs. Astrocytes were stained with GFAP (gray), and APDAs were recognized by p62 (blue) accumulation. Scale bar, 10 μm. b, Quantification of the number of inhibitory synapses around normal astrocytes and APDAs. (n = 3 mice, dots on the bar graph indicate data obtained from individual ROIs). c, Quantification of the number of excitatory synapses around normal astrocytes and APDAs in the hippocampi of 9-month-old mice injected with rapamycin and CQ (n = 3 mice, dots on the bar graph indicate data obtained from individual ROIs). b,c multiple t-tests followed by Two-stage linear step-up procedure of Benjamini, Krieger and Yekutieli. The P-values are indicated in the source data file (b,c). Mean ± s.e.m.
Extended Data Fig. 7 Effects of AD APDAs on neighboring synapses in APP/PS1 mice.
a,b Representative confocal z-stack images of APDAs in 18-month-old APP/PS1 hippocampi stained for CLDN10 (a, red), GSTM1 (b, red) and s100β (green). Scale bar, 10 μm. c, Representative confocal z-stack images of normal astrocytes and APDAs (s100β, green) stained with p62 (blue) and LC3B (red) in the hippocampi of 18-month-old APP/PS1 mice. Scale bar, 10 μm. d, Representative confocal z-stack images of the 18-month-old WT (left panels) and APP/PS1 (right panels) hippocampi stained for s100β (green), SCRG1 (red) and 6E10 (blue). Scale bar, 50 μm. e, Violin plots showing the expression of Scrg1, Gstm1 and Cldn10 in astrocyte clusters from 2-year-old APP/PS1 mice. f, Representative electron microscopy images showing nucleus inside an AD APDA (light blue). The lower panel of the enlarged image of nucleus show neither apoptotic nor necrotic feature such as large chromatin clump or nuclear condensation/fragmentation. g, Representative confocal z-stack images of dendrites (EYFP, green) in contact with the fine processes of astrocytes (upper panel) and swollen processes of APDAs (lower panel) from 18-month-old Thy1-EYFP-H; APP/PS1 mice. The processes of astrocytes were labeled with s100β (red), and swollen processes were labeled with p62 (blue). Scale bar, 10 μm. h, Quantification of the number of dendritic spines normalized to dendrite length (n = 6 mice, dots on the bar graph indicate data obtained from individual ROIs). h, two-tailed Student’s t-test. ***P = 0.0001 Mean ± s.e.m.
Extended Data Fig. 8 Ultra-fine 3D structures of astrocytic processes from normal astrocytes and APDAs.
a,b, Representative electron microscopy images showing processes of normal astrocyte (a, green) and APDA (b, red) from 18-month-old APP/PS1 hippocampi. Insets show magnified views of astrocytic processes. Normal astrocytic processes show close physical association with synapses (a, red circles), whereas APDA’s vacuolated processes fail to form contacts with synapses (b). Scale bar, 10 μm.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Rights and permissions
About this article
Cite this article
Lee, E., Jung, YJ., Park, Y.R. et al. A distinct astrocyte subtype in the aging mouse brain characterized by impaired protein homeostasis. Nat Aging 2, 726–741 (2022). https://doi.org/10.1038/s43587-022-00257-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-022-00257-1
This article is cited by
-
Distinct forebrain regions define a dichotomous astrocytic profile in multiple system atrophy
Acta Neuropathologica Communications (2024)
-
Estrogen receptor beta in astrocytes modulates cognitive function in mid-age female mice
Nature Communications (2023)
-
Contribution of Glial Cells to Polyglutamine Diseases: Observations from Patients and Mouse Models
Neurotherapeutics (2023)
-
Immortalized Alzheimer’s Disease Astrocytes: Characterization of Their Proteolytic Systems
Molecular Neurobiology (2023)
-
Astrocytic traffic jams in the aging brain
Nature Aging (2022)