Abstract
An increasing number of identified Parkinson’s disease (PD) risk loci contain genes highly expressed in innate immune cells, yet their role in pathology is not understood. We hypothesized that PD susceptibility genes modulate disease risk by influencing gene expression within immune cells. To address this, we generated transcriptomic profiles of monocytes from healthy subjects and 230 individuals with sporadic PD. We observed dysregulation of mitochondrial and proteasomal pathways. We also generated transcriptomic profiles of primary microglia from brains of 55 subjects and observed discordant transcriptomic signatures of mitochondrial genes in PD monocytes and microglia. We further identified 17 PD susceptibility genes whose expression, relative to each risk allele, was altered in monocytes. These findings reveal widespread transcriptomic alterations in PD monocytes, with some being distinct from microglia, and facilitate efforts to understand the roles of myeloid cells in PD as well as the development of biomarkers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Processed read counts and full eQTL summary statistics are available from the Zenodo online data sharing portal at https://zenodo.org/record/4715907. Raw RNA-seq data and genotypes from the MyND cohort have been made available via dbGAP (study accession ID: phs002400.v1.p1) at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002400.v1.p1. RNA-seq data and genotypes of the PPMI cohort were obtained from the AMP-PD knowledge platform. For up-to-date information on the study, visit https://www.amp-pd.org.
Code availability
The code used for the primary analysis is available on GitHub at https://github.com/RajLabMSSM/MyND-Analysis. Any additional code used for analysis is available upon request from the corresponding author.
References
Poewe, W. et al. Parkinson disease. Nat. Rev. Dis. Primers 3, 17013 (2017).
Ohnmacht, J., May, P., Sinkkonen, L. & Krüger, R. Missing heritability in Parkinson’s disease: the emerging role of non-coding genetic variation. J. Neural Transm. 127, 729–748 (2020).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
Li, Y. I., Wong, G., Humphrey, J. & Raj, T. Prioritizing Parkinson’s disease genes using population-scale transcriptomic data. Nat. Commun. 10, 994 (2019).
Gagliano, S. A. et al. Genomics implicates adaptive and innate immunity in Alzheimer’s and Parkinson’s diseases. Ann. Clin. Transl. Neurol. 3, 924–933 (2016).
Raj, T. et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523 (2014).
Reynolds, R. H. et al. Moving beyond neurons: the role of cell type-specific gene regulation in Parkinson’s disease heritability. NPJ Parkinsons Dis. 5, 6 (2019).
Bae, E.-J. et al. Antibody-aided clearance of extracellular α-synuclein prevents cell-to-cell aggregate transmission. J. Neurosci. 32, 13454–13469 (2012).
Choi, I. et al. Microglia clear neuron-released α-synuclein via selective autophagy and prevent neurodegeneration. Nat. Commun. 11, 1386 (2020).
Duffy, M. F. et al. Correction to: Lewy body-like alpha-synuclein inclusions trigger reactive microgliosis prior to nigral degeneration. J. Neuroinflammation 15, 129 (2018).
Fellner, L. et al. Toll-like receptor 4 is required for α-synuclein dependent activation of microglia and astroglia. Glia 61, 349–360 (2013).
Doorn, K. J. et al. Microglial phenotypes and Toll-like receptor 2 in the substantia nigra and hippocampus of incidental Lewy body disease cases and Parkinson’s disease patients. Acta Neuropathol. Commun. 2, 90 (2014).
Olanow, C. W. et al. Temporal evolution of microglia and α-synuclein accumulation following foetal grafting in Parkinson’s disease. Brain 142, 1690–1700 (2019).
Grozdanov, V. et al. Increased immune activation by pathologic α‐synuclein in Parkinson’s disease. Ann. Neurol. 86, 593–606 (2019).
Herz, J., Filiano, A. J., Smith, A., Yogev, N. & Kipnis, J. Myeloid cells in the central nervous system. Immunity 46, 943–956 (2017).
Harms, A. S. et al. α-Synuclein fibrils recruit peripheral immune cells in the rat brain prior to neurodegeneration. Acta Neuropathol. Commun. 5, 85 (2017).
Harms, A. S. et al. Peripheral monocyte entry is required for alpha-synuclein induced inflammation and neurodegeneration in a model of Parkinson disease. Exp. Neurol. 300, 179–187 (2018).
Bliederhaeuser, C. et al. Age-dependent defects of alpha-synuclein oligomer uptake in microglia and monocytes. Acta Neuropathol. 131, 379–391 (2016).
Wijeyekoon, R. S. et al. Monocyte function in Parkinson’s disease and the impact of autologous serum on phagocytosis. Front. Neurol. 9, 870 (2018).
Braak, H. et al. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 24, 197–211 (2003).
Scheperjans, F., Derkinderen, P. & Borghammer, P. The gut and Parkinson’s disease: hype or hope? J. Parkinsons Dis. 8, S31–S39 (2018).
Chapelet, G., Leclair-Visonneau, L., Clairembault, T., Neunlist, M. & Derkinderen, P. Can the gut be the missing piece in uncovering PD pathogenesis? Parkinsonism Relat. Disord. 59, 26–31 (2019).
Klingelhoefer, L. & Reichmann, H. Pathogenesis of Parkinson disease—the gut–brain axis and environmental factors. Nat. Rev. Neurol. 11, 625–636 (2015).
Schlachetzki, J. C. M. et al. A monocyte gene expression signature in the early clinical course of Parkinson’s disease. Sci. Rep. 8, 10757 (2018).
Donega, V. et al. Transcriptome and proteome profiling of neural stem cells from the human subventricular zone in Parkinson’s disease. Acta Neuropathol. Commun. 7, 84 (2019).
Tan, E.-K. et al. Parkinson disease and the immune system—associations, mechanisms and therapeutics. Nat. Rev. Neurol. 16, 303–318 (2020).
Hoehn, M. M. & Yahr, M. D. Parkinsonism: onset, progression and mortality. Neurology 17, 427–442 (1967).
Cuperfain, A. B., Zhang, Z. L., Kennedy, J. L. & Gonçalves, V. F. The complex interaction of mitochondrial genetics and mitochondrial pathways in psychiatric disease. Mol. Neuropsychiatry 4, 52–69 (2018).
Roth, J., Vogl, T., Sorg, C. & Sunderkötter, C. Phagocyte-specific S100 proteins: a novel group of proinflammatory molecules. Trends Immunol. 24, 155–158 (2003).
Xia, C., Braunstein, Z., Toomey, A. C., Zhong, J. & Rao, X. S100 proteins as an important regulator of macrophage inflammation. Front. Immunol. 8, 1908 (2017).
Sathe, K. et al. S100B is increased in Parkinson’s disease and ablation protects against MPTP-induced toxicity through the RAGE and TNF-α pathway. Brain 135, 3336–3347 (2012).
Jinn, S. et al. TMEM175 deficiency impairs lysosomal and mitochondrial function and increases α-synuclein aggregation. Proc. Natl Acad. Sci. USA 114, 2389–2394 (2017).
Maiese, K., Chong, Z. Z., Shang, Y. C. & Wang, S. mTOR: on target for novel therapeutic strategies in the nervous system. Trends Mol. Med. 19, 51–60 (2013).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Gordon, S. & Taylor, P. R. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5, 953–964 (2005).
Zawada, A. M. et al. SuperSAGE evidence for CD14++CD16+ monocytes as a third monocyte subset. Blood 118, e50–e61 (2011).
Mukherjee, R. et al. Non-classical monocytes display inflammatory features: validation in sepsis and systemic lupus erythematous. Sci. Rep. 5, 13886 (2015).
Grozdanov, V. et al. Inflammatory dysregulation of blood monocytes in Parkinson’s disease patients. Acta Neuropathol. 128, 651–663 (2014).
Smith, A. M. et al. Mitochondrial dysfunction and increased glycolysis in prodromal and early Parkinson’s blood cells. Mov. Disord. 33, 1580–1590 (2018).
Hoffman, G. E. & Roussos, P. Dream: powerful differential expression analysis for repeated measures designs. Bioinformatics. 37, 192–201 (2021).
Smajić, S. et al. Single-cell sequencing of the human midbrain reveals glial activation and a neuronal state specific to Parkinson’s disease. Preprint at https://www.medrxiv.org/content/10.1101/2020.09.28.20202812v1 (2020).
Wang, Q. et al. The landscape of multiscale transcriptomic networks and key regulators in Parkinson’s disease. Nat. Commun. 10, 5234 (2019).
Yao, D. W., O’Connor, L. J., Price, A. L. & Gusev, A. Quantifying genetic effects on disease mediated by assayed gene expression levels. Nat. Genet. 52, 626–633 (2020).
Young, A., Kumasaka, N., Calvert, F. & Hammond, T. R. A map of transcriptional heterogeneity and regulatory variation in human microglia. Nat. Genet. 53, 861–868 (2021).
Ng, B. et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat. Neurosci. 20, 1418–1426 (2017).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Nott, A. et al. Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science 366, 1134–1139 (2019).
Chattaragada, M. S. et al. FAM49B, a novel regulator of mitochondrial function and integrity that suppresses tumor metastasis. Oncogene 37, 697–709 (2018).
Schapira, A. H. et al. Mitochondrial complex I deficiency in Parkinson’s disease. J. Neurochem. 54, 823–827 (1990).
Mizuno, Y. et al. Deficiencies in Complex I subunits of the respiratory chain in Parkinson’s disease. Biochem. Biophys. Res. Commun. 163, 1450–1455 (1989).
Teves, J. M. Y. et al. Parkinson’s disease skin fibroblasts display signature alterations in growth, redox homeostasis, mitochondrial function, and autophagy. Front. Neurosci. 11, 737 (2017).
Ryan, B. J., Hoek, S., Fon, E. A. & Wade-Martins, R. Mitochondrial dysfunction and mitophagy in Parkinson’s: from familial to sporadic disease. Trends Biochem. Sci. 40, 200–210 (2015).
Seibler, P. et al. Mitochondrial Parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J. Neurosci. 31, 5970–5976 (2011).
Mortiboys, H. et al. Mitochondrial function and morphology are impaired in parkin-mutant fibroblasts. Ann. Neurol. 64, 555–565 (2008).
Mortiboys, H., Johansen, K. K., Aasly, J. O. & Bandmann, O. Mitochondrial impairment in patients with Parkinson disease with the G2019S mutation in LRRK2. Neurology 75, 2017–2020 (2010).
Annesley, S. J. et al. Immortalized Parkinson’s disease lymphocytes have enhanced mitochondrial respiratory activity. Dis. Model. Mech. 9, 1295–1305 (2016).
Parnetti, L. et al. CSF and blood biomarkers for Parkinson’s disease. Lancet Neurol. 18, 573–586 (2019).
Parnetti, L. et al. Cerebrospinal fluid biomarkers in Parkinson disease. Nat. Rev. Neurol. 9, 131–140 (2013).
Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014).
Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019).
Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014).
Wood, A. R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014).
Weissbrod, O., Hormozdiari, F., Benner, C. & Cui, R. Functionally-informed fine-mapping and polygenic localization of complex trait heritability. Nat. Genet. 52, 1355–1363 (2019).
Nextflow – a DSL for parallel and scalable computational pipelines. https://www.nextflow.io/ (accessed 23 August 2021).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
GENCODE – Human Release 30. https://www.gencodegenes.org/human/release_30.html (accessed 23 August 2021).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Feng, Y.-Y. et al. RegTools: integrated analysis of genomic and transcriptomic data for discovery of splicing variants in cancer. Preprint at https://www.biorxiv.org/content/10.1101/436634v2 (2018).
Babraham Bioinformatics – FastQC. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed 23 August 2021).
Picard. https://broadinstitute.github.io/picard/ (accessed 23 August 2021).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Hoffman, G. E. & Schadt, E. E. variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics. 17, 483 (2016).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Gomes, A. V. Genetics of proteasome diseases. Scientifica 2013, 637629 (2013).
Buja, A. & Eyuboglu, N. Remarks on parallel analysis. Multivariate Behav. Res. 27, 509–540 (1992).
Parsana, P. et al. Addressing confounding artifacts in reconstruction of gene co-expression networks. Genome Biol. 20, 94 (2019).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Pedersen, T. L. ggraph. GitHub https://github.com/thomasp85/ggraph (2017).
Stoeckius, M. et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Durinck, S. et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 (2005).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2018).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426#:~:text=UMAP%20(Uniform%20Manifold%20Approximation%20and,applies%20to%20real%20world%20data (2018).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 (1995).
Prakash, K. G. et al. Neuroanatomical changes in Parkinson’s disease in relation to cognition: an update. J. Adv. Pharm. Technol. Res. 7, 123–126 (2016).
Goldman, J. G. et al. Corpus callosal atrophy and associations with cognitive impairment in Parkinson disease. Neurology 88, 1265–1272 (2017).
Kendi, A. T. K., Lehericy, S. & Luciana, M. Altered diffusion in the frontal lobe in Parkinson disease. Am. J. Neororadiol. 29, 501–505 (2008).
Wayne Martin, W. R., Wieler, M., Gee, M. & Camicioli, R. Temporal lobe changes in early, untreated Parkinson’s disease. Mov. Disord. 24, 1949–1954 (2009).
Melief, J. et al. Characterizing primary human microglia: a comparative study with myeloid subsets and culture models. Glia 64, 1857–1868 (2016).
gtex-pipeline. (Github). https://github.com/broadinstitute/gtex-pipeline
Stegle, O., Parts, L., Piipari, M., Winn, J. & Durbin, R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 7, 500–507 (2012).
Delaneau, O. et al. A complete tool set for molecular QTL discovery and analysis. Nat. Commun. 8, 15452 (2017).
Fairfax, B. P. et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014).
Aguet, F. et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
summary-gwas-imputation. (Github). https://github.com/hakyimlab/summary-gwas-imputation
HaploReg v.4.1. https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php (accessed 23 August 2021).
Schilder, B. M., Humphrey, J. & Raj, T. echolocatoR: an automated end-to-end statistical and functional genomic fine-mapping pipeline. Preprint at https://www.biorxiv.org/content/10.1101/2020.10.22.351221v1 (2020).
Wang, G., Sarkar, A., Carbonetto, P. & Stephens, M. A simple new approach to variable selection in regression, with application to genetic fine-mapping. https://rss.onlinelibrary.wiley.com/doi/10.1111/rssb.12388 (2020).
Gazal, S. et al. Linkage disequilibrium–dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 49, 1421–1427 (2017).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Gazal, S. et al. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat. Genet. 50, 1600–1607 (2018).
Acknowledgements
We thank the study participants for providing blood samples and for their generous gifts of brain donation to the MyND study. We thank the Netherlands Brain Bank and the Neuropathology Brain Bank & Research Core at Mount Sinai for assistance in collecting human brain samples. We thank the Flow Cytometry Core and the Human Immune Monitoring Center at Icahn School of Medicine at Mount Sinai (ISMMS) for optimization of cell isolations; Genewiz Inc. for RNA-seq; C. Proukakis for feedback on the manuscript; S. Kim-Schulze for help in optimizing PBMC and monocyte isolation; Y.-C. Wang for help with processing of RNA-seq data; J. Fernandez-Lopez for help with data analysis; members of the Ronald Loeb Center for AD for helpful discussions; and research participants and employees of 23andMe who contributed to PD GWAS. Data used in the preparation of this article were obtained from the AMP-PD Knowledge Platform. AMP-PD—a public–private partnership—is managed by the US National Institutes of Health (NIH) and funded by Celgene, GSK, the Michael J. Fox Foundation for Parkinson’s Research, the National Institute of Neurological Disorders and Stroke, Pfizer, Sanofi and Verily. PPMI—a public–private partnership—is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners (names of all PPMI funding partners can be found at www.ppmi-info.org/fundingpartners). The PPMI Investigators did not participate in reviewing the data analysis or content of the manuscript. For up-to-date information on the study, visit www.ppmi-info.org. This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the ISMMS. T.R. was supported by grants from the Michael J. Fox Foundation (nos. 4899 and 16743) and NIH (nos. NINDS R01-NS116006, NINDS U01-NS120256, NIA R01-AG054005, NIA R21-AG063130 and NIA U01 P50-AG005138). R.S-P. is supported by grants from the NIH (nos. NINDS U01-NS107016 and NINDS U01-NS094148-01) and Bigglesworth Family Foundation. K.F. is supported by NIH (no. F32 AG056098). E.N. is supported by a fellowship from the Ramon Areces Foundation (Spain). The research reported in this paper was additionally supported by the Office of Research Infrastructure of the NIH under award nos. S10OD018522 and S10OD026880.
Author information
Authors and Affiliations
Contributions
T.R. conceived the study. T.R., E.N. and E.U. led the project, designed and performed the experiments and analysis and wrote the manuscript. K.d.P.L. performed coexpression network and microglial transcriptomic analysis. M.P. and E.N. optimized the experimental approach. M.P., M.Z. and A.A. performed experimental isolations. B.M.S. and J.H. performed single-cell and splicing analyses, respectively. T.S., C.A., B.H. and S.S. contributed to clinical coordination. R.A.V. contributed to data analyses. G.R. and S.F. were responsible for the implementation and recruitment of patients with movement disorder at the Fresco Institute for Parkinson’s and Movement Disorders at New York University (NYUMD) and Bendheim Parkinson and Movement Disorders Center at Mount Sinai (BPMD). M.J.C. contributed to genetic and population characterization. D.R., S.E., R.A.O., V.S., M.Swan., S.B. and R.S.-P. contributed to recruitment and clinical characterization of patients from Movement Disorder Center at Mount Sinai Beth Israel (MSBI). C.W.Z. and M.Sano. contributed to recruitment of donors from the Alzheimer’s Research Center (ADRC). A.C.P. implemented recruitment of donors from the Center for Cognitive Health (CCH). R.R., R.H.W. and W.T. helped with recruitment of patients from BPMD. T.A. and A.M.G. helped with intellectual discussion and interpretation of the results. K.F., R.H.W. and J.F.C. contributed to brain sample collection. G.J.L.S. and L.d.W. contributed to microglial isolation and characterization. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
A.M.G. served on the scientific advisory board for Denali Therapeutics from 2015 to 2018 and has served as a consultant to AbbVie, Biogen, Eisai, Illumina and GSK. R.S.-P. and S.B. have served as consultants to Denali Therapeutics. R.H.W. has served as consultant to Neurocine Biosciences, Inc. The other authors declare no competing interests.
Additional information
Peer review information Nature Aging thanks David Craig, Richard Youle and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Experimental flow outline and demographic/clinical information for subjects for monocytes isolation.
(A) Blood was collected from five independent clinics across New York City (ADRC, CCH, MSMD, BIMD, and NYUMD; details described in Methods) and transferred to the Icahn School of Medicine at Mount Sinai for monocyte sorting and RNA/DNA isolation. Samples were genotyped for common SNPs using Global Screening Array (GSA) and LRRK2, GBA and APOE were independently genotyped. RNA-seq was performed at Genewiz Inc. in three independent and randomized batches. DNA and RNA data was subjected to stringent QC, DNA data was imputed and ancestry was calculated. DNA and RNA were compared to the identification of miss-matches prior outlier identification. After QC, a total of 230 samples were used for subsequent analysis. (B) Demographic, (C) genotype and (D) clinical variables describing the 230 samples included in the study.
Extended Data Fig. 2 Differential expression analysis at the transcript level in PD and controls derived monocytes.
(A) MA plot showing the fold-change (log2 scale) at the transcript level in the y-axis and the mean of log2counts (x-axis), highlighting the DETs at FDR < 0.05 in red. (B) Volcano plot showing the fold-change (log2 scale) of transcripts between PD-monocytes (n = 135) and controls (n = 95) (x-axis) and their significance in the y-axis -log10 P-value scale). DETs at FDR < 0.05 are highlighted in red (upregulated) and blue (downregulated). Moderated t-statistic (two sided) is used for statistical test (see R package limma). (C) Pathway enrichment analysis for the upregulated (n = 230 independent samples) and (D) downregulated DETs using Biological processes from GSEA. Significance is represented in the x-axis (-log10 P-value scale of the q-value). Only the 20 most significant pathways (q-value < 0.05) with a minimum overlap of 5 genes are shown. Pathways are grouped and colored by biological related processes. n = 230 independent samples.
Extended Data Fig. 3 Differential expression analysis at the splicing level in PD and controls derived monocytes.
(A) Histogram reflecting the counts (y-axis) and the % of missingness (x-axis). (B) Volcano plot showing the delta PSI of genes with splicing events in PD-monocytes (n = 135) and controls (n = 95) (x-axis) and their significance in the y-axis (-log10 P-value scale). DSs at FDR < 0.05 are highlighted in red (delta PSI > 0) and blue (delta PSI < 0). Positive delta-PSI indicates that the long isoform is favored whereas negative delta-PSI indicates preference for the short isoform. Chi-squared is used for statistical test (C) Pathway enrichment analysis for the DSs at FDR < 0.0.5 (left panel) DSs + DEGs at FDR < 0.05 (right panel) using Biological processes from GSEA. Significance is represented in the x-axis (-log10 P-value scale of the q-value). Only the 20 most significant pathways (q-value < 0.05) with a minimum overlap of 5 genes are shown. Pathways are grouped and colored by biological related processes. n = 230 independent samples. (D) Examples of genes showing significant splicing events.
Extended Data Fig. 4 Module enrichment for biological pathways.
(A) Enrichment of modules (x-axis) containing co-expressed genes for specific biological pathways and curated gene sets (y-axis). Modules are represented by color names and are ordered by size. Enrichment for selected gene sets and GO biological processes (top panel). The size and color of the circles indicate the significance level (-log10 P-value). Enrichments for PD heritability, using stratified LD score regression (bottom panel). The size and color of circles indicate the enrichment value (from LD score) and significance level (-log10 P-value) of enrichment, respectively. Only modules that were significant at a nominal P-value < 0.05 are shown here. (B) Barchart showing Pearson correlation coefficient (r) (x-axis) of three modules (y-axis) significantly associated with PD (FDR < 0.05) determined by the module eigengene analysis. Numbers on the plot represent adjusted P-values, Two-sided Wilcoxon rank-signed test. n = 230 independent samples.
Extended Data Fig. 5 Monocyte subcluster characterization by single-cell analysis.
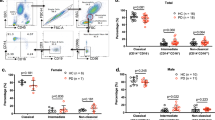
(A) Proportions of the 3 main monocyte sub-clusters using FACS (n = 11 controls and 11 PD). No statistical differences were obtained between groups. (B) Cell proportions of the 6 sub-clusters obtained by unsupervised clustering with monocle3 in scRNA-seq (n = 3 controls and 7 PD). No statistical differences were obtained between cases and controls in cell proportions. Cluster 1 corresponds to classical monocytes and cluster 2 to intermediate monocytes. (C) Top: UMAP colored by diagnosis (green = controls, yellow = PD). Bottom: UMAP colored by CD14 and FCGR3A (CD16) marker genes expression. (D) Histogram showing the variance (y-axis) explained by the 20 first PCA components (x-axis). (E) Histogram showing the frequency of the genes colored by diagnosis (green = control, yellow = PD). (F) Expression of mitochondrial genes by each cluster and divided by diagnosis. Boxplots: the line represents the median. The boxes extend from the 25th - 75th percentile and the lines extend 1.5 times the interquartile range. n = 10 independent donors.
Extended Data Fig. 6 Fresh microglia transcriptome analysis.
Microglia transcriptomic profiling was performed from 22 samples from 13 PD donors and 106 samples from 42 control donors. (A) Experimental workflow for the generation of human microglial transcriptomic profiles (B) Tables describing the samples included in the study (top: demographic information, middle: clinical information, bottom: brain regions). CC: Corpus Callosum; MFG: medial frontal gyrus; STG: Superior temporal gyrus; THA: thalamus; SVZ: subventricular zone; SN: substantia nigra (C) Heatmap for the expression of marker genes of different brain cell types (red: microglia, dark blue: astrocytes, green: neurons, light blue: oligodendrocytes). (D) Microglial isolation purity assessed by qPCR comparing the brain homogenate, and the positive and negative fractions after CD11b magnetic beads comparing microglial markers (P2RY12, CXCR1, TREM2) and astrocytic markers (GFAP, FGFR3). (E) Violin plot showing the % of variance (y-axis) explained by known covariates (x-axis) by variancePartition. Each dot represents a gene. (F) PCA after regressing out covariates colored by diagnosis (left panel), brain region (middle panel), postmortem interval (right panel). n = 128 samples from 55 independent donors. (G) Volcano plot showing the fold-change of genes (log2 scale) between PD-microglia (22 samples from 13 donors) and controls (106 samples from 42 donors) (x-axis) and their significance (y-axis, -log10 scale). Moderated t-statistic (two sided) is used for statistical test (see R package DREAM). (H) Expression of selected mitochondria-specific genes in microglia. Adjusted gene expression levels after normalization are shown. Boxplots: the line represents the median. The boxes extend from the 25th - 75th percentile and the lines extend 1.5 times the interquartile range. n = 128 samples from 55 independent donors.
Supplementary information
Rights and permissions
About this article
Cite this article
Navarro, E., Udine, E., Lopes, K.d.P. et al. Dysregulation of mitochondrial and proteolysosomal genes in Parkinson’s disease myeloid cells. Nat Aging 1, 850–863 (2021). https://doi.org/10.1038/s43587-021-00110-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-021-00110-x
This article is cited by
-
The contribution of the peripheral immune system to neurodegeneration
Nature Neuroscience (2023)
-
Epigenome-wide association study of peripheral immune cell populations in Parkinson’s disease
npj Parkinson's Disease (2023)
-
Human iPSC-derived microglia carrying the LRRK2-G2019S mutation show a Parkinson’s disease related transcriptional profile and function
Scientific Reports (2023)
-
Transcriptome deregulation of peripheral monocytes and whole blood in GBA-related Parkinson’s disease
Molecular Neurodegeneration (2022)
-
Genetic analysis of the human microglial transcriptome across brain regions, aging and disease pathologies
Nature Genetics (2022)