Abstract
Like many adult stem cell populations, the capacity of oligodendrocyte progenitor cells (OPCs) to proliferate and differentiate is substantially impaired with aging. Previous work has shown that tissue-wide transient expression of the pluripotency factors Oct4, Sox2, Klf4 and c-Myc extends lifespan and enhances somatic cell function. Here we show that just one of these factors, c-Myc, is sufficient to determine the age state of OPC: c-Myc expression in aged OPCs drives their functional rejuvenation, while its inhibition in neonatal OPCs induces an aged-like phenotype, as determined by in vitro assays and transcriptome analysis. Increasing c-Myc expression in aged OPCs in vivo restores their proliferation and differentiation capacity, thereby enhancing regeneration in an aged central nervous system environment. Our results directly link Myc to cellular activity and cell age state, with implications for understanding regeneration in the context of aging, and provide important insights into the biology of stem cell aging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The RNA-seq data of neonatal and aged OPCs generated during this study are available at SRA: PRJNA756964. Other data supporting the findings of the study can be requested from the corresponding author.
Code availability
No new code for the analysis of data was created.
References
Zawadzka, M. et al. CNS-resident glial progenitor/stem cells produce Schwann cells as well as oligodendrocytes during repair of CNS demyelination. Cell Stem Cell 6, 578–590 (2010).
Hill, R. A., Li, A. M. & Grutzendler, J. Lifelong cortical myelin plasticity and age-related degeneration in the live mammalian brain. Nat. Neurosci. 21, 683–695 (2018).
Young, K. M. et al. Oligodendrocyte dynamics in the healthy adult CNS: evidence for myelin remodeling. Neuron 77, 873–885 (2013).
Neumann, B. et al. Metformin restores CNS remyelination capacity by rejuvenating aged stem cells. Cell Stem Cell 25, 473–485 (2019).
Segel, M. et al. Niche stiffness underlies the ageing of central nervous system progenitor cells. Nature 573, 130–134 (2019).
Sim, F. J., Zhao, C., Penderis, J. & Franklin, R. J. M. The age-related decrease in CNS remyelination efficiency is attributable to an impairment of both oligodendrocyte progenitor recruitment and differentiation. J. Neurosci. 22, 2451–2459 (2002).
Lapasset, L. et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 25, 2248–2253 (2011).
Ocampo, A. et al. In vivo amelioration of age-associated hallmarks by partial reprogramming. Cell 167, 1719–1733 (2016).
Sarkar, T. J. et al. Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat. Commun. 11, 1545 (2020).
Tabula Muris Consortium. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020).
Magri, L. et al. c-Myc-dependent transcriptional regulation of cell cycle and nucleosomal histones during oligodendrocyte differentiation. Neuroscience 276, 72–86 (2014).
Zirath, H. et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl Acad. Sci. USA 110, 10258–10263 (2013).
Scognamiglio, R. et al. Myc depletion induces a pluripotent dormant state mimicking diapause. Cell 164, 668–680 (2016).
Tamamori-Adachi, M. et al. DNA damage response induced by Etoposide promotes steroidogenesis via GADD45A in cultured adrenal cells. Sci. Rep. 8, 9636 (2018).
Moyon, S. et al. Demyelination causes adult CNS progenitors to revert to an immature state and express immune cues that support their migration. J. Neurosci. 35, 4–20 (2015).
Challis, R. C. et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 14, 379–414 (2019).
Suzuki, K. et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149 (2016).
Cosgrove, B. D. et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med. 20, 255–264 (2014).
Kalamakis, G. et al. Quiescence modulates stem cell maintenance and regenerative capacity in the aging brain. Cell 176, 1407–1419 (2019).
Wilson, A. et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 18, 2747–2763 (2004).
Ryall, J. G., Cliff, T., Dalton, S. & Sartorelli, V. Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell 17, 651–662 (2015).
Cerletti, M., Jang, Y. C., Finley, L. W. S., Haigis, M. C. & Wagers, A. J. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell 10, 515–519 (2012).
Mihaylova, M. M., Sabatini, D. M. & Yilmaz, O. H. Dietary and metabolic control of stem cell function in physiology and cancer. Cell Stem Cell 14, 292–305 (2014).
Beerman, I., Seita, J., Inlay, M. A., Weissman, I. L. & Rossi, D. J. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 15, 37–50 (2014).
Basak, O. et al. Induced quiescence of Lgr5+ stem cells in intestinal organoids enables differentiation of hormone-producing enteroendocrine cells. Cell Stem Cell 20, 177–190 (2017).
Leeman, D. S. et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283 (2018).
Laurenti, E. et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3, 611–624 (2008).
Chiariello, M., Marinissen, M. J. & Gutkind, J. S. Regulation of c-myc expression by PDGF through Rho GTPases. Nat. Cell Biol. 3, 580–586 (2001).
Woodruff, R. H., Fruttiger, M., Richardson, W. D. & Franklin, R. J. M. Platelet-derived growth factor regulates oligodendrocyte progenitor numbers in adult CNS and their response following CNS demyelination. Mol. Cell. Neurosci. 25, 252–262 (2004).
Vervoorts, J., Lüscher-Firzlaff, J. & Lüscher, B. The ins and outs of MYC regulation by posttranslational mechanisms. J. Biol. Chem. 281, 34725–34729 (2006).
Ge, Y. et al. Stem cell lineage infidelity drives wound repair and cancer. Cell 169, 636–650 (2017).
Kotter, M. R., Li, W.-W., Zhao, C. & Franklin, R. J. M. Myelin impairs CNS remyelination by inhibiting oligodendrocyte precursor cell differentiation. J. Neurosci. 26, 328–332 (2006).
McMurran, C. E., Zhao, C. & Franklin, R. J. M. Toxin-based models to investigate demyelination and remyelination. Methods Mol. Biol. 1936, 377–396 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Olive, P. L. & Banáth, J. P. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 1, 23–29 (2006).
Collins, A. R. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 26, 249–261 (2004).
Ho, J. W. K., Stefani, M., dos Remedios, C. G. & Charleston, M. A. Differential variability analysis of gene expression and its application to human diseases. Bioinformatics 24, i390–i398 (2008).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Acknowledgements
This work was supported by funding from the UK Multiple Sclerosis Society (MS50), MedImmune, the Adelson Medical Research Foundation and by a core support grant from the Wellcome Trust and MRC to the Wellcome Trust–Medical Research Council Cambridge Stem Cell Institute (no. 203151/Z/16/Z). B.N. was supported by a Kim and Julianna Silverman research fellowship and a donation from the Mikheelev family trust. K.S.R. was supported by a postdoctoral fellowship from the Multiple Sclerosis Society of Canada. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization was carried out by B.N., M.S. and R.J.M.F. Methodology was the responsibility of M.S. and B.N. Investigation was performed by M.S., B.N., C.Z., P.T., A.Y., S.F., A.S., C.Z.-Y.C. and J.F.C. Formal analysis was conducted by M.S., B.N., C.Z., K.S.R., K.J.C. and R.J.F.M. B.N., M.S., K.J.C. and R.J.M.F. wrote the article. T.G. performed microarray analysis. R.J.M.F. sourced funding acquisition. K.J.C. and R.J.M.F. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Aging thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Transfection of Myc but not Sox2 reactivates aged rat OPCs.
Schematic illustrating the transfection strategy using mRNAs. (b,c), Representative images and quantifications of the transfection efficiency of modified mRNA in aged OPCs. Control transfections were transfected with lipofectamine without mRNA encoding GFP. Scale bars represent 100 µm. N = 3 biological repeats. (d), Scatterplots of scRNAseq data for c-MYC expression in Olig2 and CSPG4 expressing cells in 3 and 24 month old animals. Data produced from (https://tabula-muris-senis.ds.czbiohub.org). (e), qPCR results measuring relative abundance of MYC mRNA in freshly MACs sorted neonatal and aged OPCs. N = 3 biological repeats. (f), Representative images of EdU incorporation in aged OPCs transfected with GFP or Sox2 (N = 2). (g), qPCR results measuring relative abundances of c-Myc mRNA 5d in aged OPCs after transfection with GFP or c-Myc mRNA. Means represent averages from N = 3 biological replicates. (h), qPCR results measuring relative abundances of c-Myc mRNA in proliferating or differentiating neonatal OPCs 3 days after culture in the respective culture conditions. (N = 3 biological repeats for each group). (i), Representative images of aged OPCs 5 days after transfection with c-Myc mRNA stained with the oligodendrocyte lineage markers Olig2, SOX10, and the OPC marker NG2 (n = 2 technical replicates). Unless otherwise indicated, scale bars represent 50 µm. All bar-graphs represent mean ± SD. Statistical significance was determined using a two-tailed t-test in all cases.
Extended Data Fig. 2 CRISPR mediated knockdown of c-MYC diminishes the proliferation and differentiation capacity of neonatal OPCs.
(a-b), Representative images and quantification of live cell stain CalceinAM following neonatal OPC treatment with increasing doses of Myci. (b), Quantification of cell viability as the frequency of Calcein AM/DAPI co-positive cells. N = 6 technical repeats from 3 biological repeats. (c), Schematic illustrating the strategy for the CRISPR experiments. (d), off target indel rates, predicted by CRISPOR, quantified by TIDE of the two Myc targeting gRNAs. N = 2 biological repeats. (e), Representative image of neonatal cells 5d stained with the OPC marker A2B5 and the oligodendrocyte marker MBP 5d after transfection with spCas9 and gRNAs. (f), qPCR for c-Myc in neonatal OPCs after transfection with Cas9 and control or c-Myc targeting gRNAs. (g), Representative images of EdU incorporation and MBP expression in Olig2 stained neonatal OPCs treated with either DMSO or MYC-inhibitor for 48 h and then cultured without the inhibitor or solvent for an additional 7d or 10d in proliferation or differentiation conditions respectively. (h,i), Quantification of the data for EdU incorporation and MBP expression, respectively. Scale bars represent 50 µm. Means represent N = 3 biological replicates ± SD when not specified otherwise. Statistical significance was determined using two-tailed t-tests.
Extended Data Fig. 3 Inflammatory pathways are enriched in all OPCs with an age-like phenotype.
(a), Principal component analysis (PCA) of mRNA sequencing of primary aged OPCs, primary neonatal OPCs, in vitro aged OPCs, in vitro neonatal OPCs, neonatal OPCs treated with Myci, and aged OPCs transfected with modified mRNAs encoding MYC. (b), treated Venn diagram illustrating the overlapping pathways and processes in aged OPCs, neonatal OPCs treated with Myci and neonatal OPCs cultured on stiff hydrogels (data from ref. 5). (c), NES values are plotted for example processes enriched in all ‘aged like’ OPCs. (d-e), Selected genes from various genesets show a number of differentially expressed genes between (d) aged cells transfected with GFP and c-Myc neonatal OPCs or (e) neonatal OPCs treated with DMSO or Myci. (f), Quantification of viability assay comparing neonatal and aged OPCs at the time-point of the comet assay in Fig. 3f. Means represent N = 3 biological repeats ± SD. Statistical significance was determined using a two-tailed t-test.
Extended Data Fig. 4 Increased MYC reduces DNA damage in aged OPCs while MYC inhibition impairs DNA repair in neonatal OPCs.
(a), Schematic outlining of the experiments. (b), Representative images of cells stained with the viability marker Calcein AM. (c), Quantification of the total number of cells counted in 9 microscope fields, N = 3 for 0.5, 6, 12, 25 µM ETS and N = 4 biological repeats for DMSO, 0.125, 0.25, 1, 3 µM ETS. (d), Quantification of the viability as the frequency of Calcein AM positive cells among all cells,. N = 3 biological repeats. (e), Representative images of comet assays 2 h after treatment with DMSO or 1 µM Etoposide (ETS), N = 2 biological replicates. (f), Quantification of the comet assay. (g), Representative images of comet assays of cells cultured for 48 h in DMSO or MYCi prior to exposure to ETS (2 h) and 48 in normal medium. (h), Quantification of the comet assay depicted in g. N = 2 biological replicates. ETS: Etoposide. All data are presented as mean ± SD in c,d and the mean is presented in f,h. Statistical significance was determined using a One-Way ANOVA and Dunnett’s multiple comparisons test in c, h and two-tailed t-tests for the data in d and f. Scale bars represent 50 µm.
Extended Data Fig. 5 All neonate like OPCs share pathways that reflect stem cell activity.
(a), Venn diagram illustrating the overlapping pathways and processes in neonatal OPCs, aged OPCs transfected with Myc and activated OPCs isolated from the brains of mice 5 weeks after cuprizone treatment (data from ref. 15). (b), NES values are plotted for example processes enriched in ‘neonate like’ and activated OPCs. (c), Top-5 transcription factors identified by transcription factor binding site analysis using HOMER2. Genes used for analysis were significantly higher expressed in activated versus resting OPCs (log2 fold change >1, adjusted p-value < 0.05, reanalysed data from Moyon et al., 2015). (d), Venn diagram illustrating the overlapping pathways and processes in neonatal OPCs, aged OPCs transfected with c-Myc and rejuvenated aged OPCs that were cultured on soft hydrogels (data from ref. 5). (e), NES values are plotted for example processes enriched in ‘neonate like’ and rejuvenated aged OPCs cultured on soft hydrogels.
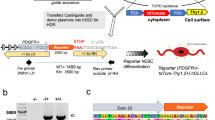
Extended Data Fig. 6 In vivo CRISPR knock-in of c-Myc into the 3’UTR of Pdgfra.
(a), Schematic overview of the predicted knock-in fragments following in vivo CRISPR in the 3’UTR of Pdgfra. The primers used to identify correct genomic integration 3 weeks following tail vein injection are highlighted as arrows. (b), Representative images of mixed glia cultures transduced with AAVs used to achieve the genetic modifications described in a. GFP is only expressed by Olig2 expressing cells. (c), Western Blot for GFP and Actin from neonatal mouse OPCs transduced with either the IRES-GFP or IRES-MYC-T2A-GFP AAVs. (N = 2 technical replicates for each experimental group). (d), PCR results confirming the integration of viral constructs into the 3’UTR of Pdgfra. (e), Representative image and of un-lesioned white/grey matter of the spinal cord reveal the efficiency of our in vivo CRISPR system and of the expression specificity of Pdgfra for Olig2 expressing OPCs—only ~10% of GFP positive cells are not Olig2 + . (f), Quantification of transduction efficiency in vivo. (g-i), Representative images of spinal cords after transduction with AAVs. Identity of GFP cells was tested for PDGFRB (pericytes), IBA1 (microglia) and GFAP (astrocytes). (j), Quantification of the data presented in g-i. N = 3 biological replicates. (k), Representative images of lesions (below the dotted line) stained for GFP, Olig2 and incorporated EdU 14d post lesion induction. Averages represent mean from N = 3 animals and scale bar represents 100 µm. (l), Quantification of the frequency of GFP positive cells among EdU + Olig2 + cells at 14 dpl. All data are presented as mean ± SD. Scale bars: b:50 µm, e: 100 µm, g-i: 25 µm, k: 100 µm.
Supplementary information
Supplementary Table 1
Complete results for GSEA comparing aged and neonatal OPCs in Fig. 2c. NES score calculations are based on a weighted Kolmogorov–Smirnov-like (K-S) statistic.
Rights and permissions
About this article
Cite this article
Neumann, B., Segel, M., Ghosh, T. et al. Myc determines the functional age state of oligodendrocyte progenitor cells. Nat Aging 1, 826–837 (2021). https://doi.org/10.1038/s43587-021-00109-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-021-00109-4
This article is cited by
-
Microglia regulation of central nervous system myelin health and regeneration
Nature Reviews Immunology (2024)
-
Restoration of neuronal progenitors by partial reprogramming in the aged neurogenic niche
Nature Aging (2024)
-
The Impact of Aging on Multiple Sclerosis
Current Neurology and Neuroscience Reports (2024)