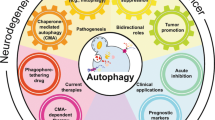
Abstract
Autophagy is a fundamental cellular process that eliminates molecules and subcellular elements, including nucleic acids, proteins, lipids and organelles, via lysosome-mediated degradation to promote homeostasis, differentiation, development and survival. While autophagy is intimately linked to health, the intricate relationship among autophagy, aging and disease remains unclear. This Review examines several emerging features of autophagy and postulates how they may be linked to aging as well as to the development and progression of disease. In addition, we discuss current preclinical evidence arguing for the use of autophagy modulators as suppressors of age-related pathologies such as neurodegenerative diseases. Finally, we highlight key questions and propose novel research avenues that will likely reveal new links between autophagy and the hallmarks of aging. Understanding the precise interplay between autophagy and the risk of age-related pathologies across organisms will eventually facilitate the development of clinical applications that promote long-term health.
Similar content being viewed by others
Main
Aging is a biological process that is characterized by time-dependent cellular and functional decline, resulting in reduced quality of life for the organism1. In line with this, aging is the primary risk factor for the development of many disorders, including cardiovascular disease (for example, stroke), cancer and neurodegenerative disease (for example, Alzheimer’s disease (AD)). Collectively, age-related ailments represent a formidable global socioeconomic burden and a significant healthcare challenge2,3. Therefore, identifying therapeutic interventions that promote ‘healthy aging’ (that is, the maintenance of functional ability in old age, enabling older individuals to independently carry out daily tasks) and simultaneously halt the progression of multiple age-related pathological conditions is of paramount importance2.
Among the many molecular changes associated with old age, altered autophagy has emerged as a feature of aging across diverse species. However, recent advances in understanding the numerous substrates of autophagy and the temporal and spatial effects of impaired autophagy regulation on tissue homeostasis have revealed a complex and multifactorial relationship between autophagy and aging. Here we examine the relationship among autophagy, aging and disease and propose novel links between specific autophagic processes and long-term tissue health, as well as possible implications for anti-aging therapeutic interventions.
Compromised autophagy is a hallmark of aging
Research over the last decade has revealed that the process of autophagy can take many different forms. Autophagy (from the Greek words auto, meaning ‘self’, and phagein, meaning ‘to eat’) is a highly conserved pathway that degrades cellular components, such as defective organelles and aggregates of misfolded protein4, through lysosomes. The process of autophagy was first described in the 1960s, but it was the identification of autophagy-related genes (ATG genes) in the 1990s that propelled major breakthroughs in unravelling the mechanistic complexities of autophagy5,6,7,8,9,10,11,12. There are three major types of autophagy: macroautophagy, microautophagy and chaperone-mediated autophagy (CMA) (Fig. 1a–c), all of which involve delivery of substrates to the lysosome for degradation (see detailed reviews in refs. 13,14). Macroautophagy (hereafter referred to as autophagy) was originally thought of as a nonselective bulk degradation process (Fig. 1a, pathway (1)). However, the discovery of selective autophagy receptors, among which p62/SQSTM1 was the first, changed this notion15,16. Today, autophagy is recognized as a highly selective cellular clearance pathway that is associated with the maintenance of cellular and tissue homeostasis17,18. Selective autophagy can be further divided into many subtypes on the basis of the specific cargos involved. These subtypes target various macromolecules (glycophagy and lipophagy) (Fig. 1a, pathways (2)–(5)), mitochondria (mitophagy) (Fig. 1a, pathway (6)), the endoplasmic reticulum (ER) (ER-phagy) (Fig. 1a, pathway (7)), parts of the nucleus (nucleophagy) (Fig. 1a, pathway (8)), pathogens (xenophagy) (Fig. 1a, pathway (9)) and lysosomes themselves (lysophagy) (Fig. 1a, pathway (10)). Below we will discuss the links among these selective autophagy pathways, aging and disease. The core process of autophagy has been described in detail elsewhere14,19. However, in brief, the core process is initiated following inhibition of mechanistic target of rapamycin (mTOR) or activation of 5′ AMP-activated protein kinase (AMPK), both of which are canonical inducers of autophagy in response to stress (for example, starvation or elevated temperatures) and physical exercise. In addition, transcription factor EB (TFEB) is an important positive regulator of autophagy and lysosomal biogenesis whose nuclear translocation is coupled to the activity of both mTOR (via phosphorylation) and AMPK (via folliculin (FLCN))20,21,22,23. Upon activation of autophagy, the process is initiated by membrane nucleation and phagophore formation followed by elongation and maturation before autophagosome fusion with the lysosome for cargo degradation and recycling. The key proteins involved in each step are presented in Fig. 2.
a, Macroautophagy (referred to herein as autophagy) (1) is a nonselective process that targets macromolecules or subcellular organelles in bulk. Cytoplasmic material is sequestered into an autophagosome and delivered to the lysosome (or endolysosome) for degradation). Selective autophagy involves recognition of specific cytoplasmic cargo via autophagy receptors that also interact with LC3 in the autophagic membrane, leading to cargo sequestration into autophagosomes that are delivered to a lysosome (or endolysosome) for degradation. This includes aggrephagy (2), where aggregated proteins are ubiquitinated and targeted by ubiquitin-binding autophagy receptors such as p62 (or NBR1); glycophagy (3), where STBD1 (genethonin-1) binds to glycogen and GABARAP, facilitating lysosomal glycogen breakdown into non-phosphorylated glucose by enzymes such as GAA; lipophagy (4), in which lysosomal lipids are degraded into free fatty acids, which are then converted into ATP; the identity of the receptor(s) (yellow) involved in sequestration of lipid droplets is unknown;; granulophagy (5), where sequestration of stress granules (RNA + protein) is mediated by Cdc48/VCP, allowing the stress granule to be delivered to the lysosome for degradation; mitophagy (6), where damaged mitochondria are bound by soluble or membrane-bound mitophagy receptors (mReceptors) that can also bind LC3, leading to engulfment of the mitochondrion into an autophagosome and subsequent delivery to a lysosome for degradation (left); in piecemeal mitophagy, degradation of parts of mitochondria occurs via binding of the outer mitochondrial membrane protein metaxin-1 (MTX1, in the extruded fraction) to LC3C, resulting in the recruitment of p62 and autophagosome formation (right); ER-phagy (7), which in mammals uses the specific receptors FAM134B, RTN3L, ATL3, SEC62, CCPG1 and TEX264, which are located in different parts of the ER; these receptors bind to LC3, leading to sequestration of the ER into an autophagosome and lysosomal degradation of the ER; nucleophagy (8), which, when triggered in mammals, results in nuclear LC3 binding to lamin B1, leading to formation of a bulge that is pinched off to the cytoplasm where degradation by autophagy occurs; xenophagy (type A, 9), where a bacterium’s DNA is detected by cGAS, a sensor that triggers a process of ubiquitination via Smurf1; this is followed by attachment of the NBR1 receptor to the ubiquitin chains and LC3 to continue the autophagy process for degradation of the bacterium; xenophagy (type B, 10), where a bacterium damages the membrane of the phagosome, exposing interior glycans that recruit galectin-8 (Gal-8), which is then recognized by NDP52 to recruit TBK1, LC3C, Nap and Sintbad; the optineurin, p62 and NDP52 receptors interact with ubiquitin on the pathogen and recruit the autophagic engulfment system, and the engulfed pathogen is then brought for degradation; and lysophagy (11), which occurs upon lysosomal membrane permeabilization and can be achieved with or without ubiquitination: recruitment of galectin-3 (Gal-3) to damaged lysosomes further recruits TRIM16 and autophagic proteins such as ULK1 and ATG16L1, and ubiquitination on the lysosome results in the recruitment of p62, which binds to LC3 to facilitate the autophagic process (left); in a parallel ubiquitin-independent process, galectin-8 is recruited to damaged lysosomes and is capable of directly binding to the NDP52 receptor that interacts with LC3 to continue the autophagic process (right). b, Microautophagy involves capture of cytoplasmic components through direct invagination of endolysosome membranes and can be nonspecific (bulk) (12) or highly specific (13,14). Examples of selective microautophagy in mammalian cells include micro-ER-phagy (13), which uses the SEC62 receptor and involves ER capture and degradation by invagination of the lysosome/endolysosome, and endosomal microautophagy of proteins with the KFERQ pentapeptide motif (14) in a process requiring the chaperone HSC70. c, CMA (15) also involves targeting of proteins containing a KFERQ pentapeptide-related motif by HSC70 and other co-chaperones such as HSP40. The substrate is then imported into the lysosome through the LAMP2A receptor for further degradation. The LAMP2A receptor is modulated by the glial fibrillary acidic protein (GFAP). Finally, in a CMA-like manner, DNAutophagy/RNAutophagy (16) can occur: nucleic acids (DNA or RNA) bind to the LAMP2C receptor (orange), which also binds to lysosomes. This process allows nucleic acids to be taken up by the lysosome. It has been proposed that a transporter called SIDT2 (green) might have a role in direct uptake of nucleic acids by the lysosome.
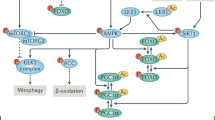
Initiation of autophagy requires the ULK1 kinase complex, which is tightly regulated by AMPK and mTOR, which act as an activator and inhibitor, respectively. AMPK activates ULK1 through phosphorylation. The ULK1 complex, composed of FIP200, ATG13 and ATG101, stimulates the class III phosphatidylinositol 3-kinase (PIK3C3) complex, which is composed of BECN1 (which can be inhibited by BCL-2), AMBRA1, ATG14L, VPS15 and VPS34. This complex then produces a pool of phosphatidylinositol 3-phosphate (PtdIns3P), which leads to the recruitment of WIPI proteins, which recover ATG9-positive vesicles from previous membranes, as well as recruiting the ATG5–ATG12–ATG16L1 (E3) complex. LC3 is first cleaved by the ATG4 protease to form cytosolic LC3-I, which is further recognized by E1 (ATG7), E2 (ATG3) and E3 components, leading to its conjugation to phosphatidylethanolamine (PE). After this process, LC3-I is referred to as LC3-II. LC3-II binds to LIR-containing autophagy receptors (AR; such as p62) bound to cargo targeted for degradation. Fusion of autophagosomes with lysosomes is mainly mediated by the assistance of RAB proteins, SNARE proteins and a HOPS complex. After fusion, the cargo is degraded by lysosomal hydrolases and the degradation products can be reused by the cell. LC3-II bound to the outer membrane is cleaved by ATG4 to be reused for a new round of lipidation.
A growing body of evidence suggests that autophagic activity declines with age in diverse organisms1. Studies in Caenorhabditis elegans, rodents and human cells have demonstrated an age-dependent reduction in lysosomal proteolytic function that thereby impairs autophagic flux24,25,26,27, exacerbating cellular impairment and contributing to the development of age-related diseases1,28,29. Further evidence stemming from Drosophila has demonstrated that aging is associated with reduced expression of several Atg genes (Atg2, Atg8a and bchs (encoding blue cheese)), which are pivotal for both autophagy initiation and activity30. In aged wild-type mice, autophagy is diminished in neuronal cells, as evidenced by decreased rates of autophagolysosomal fusion and impaired delivery of autophagy substrates to lysosomes in the hypothalamus31. Moreover, a decrease in autophagic processes was observed in brain tissue from 18- to 25-month-old mice, as demonstrated by a reduction in the levels of Atg5–Atg12 and Becn1, elevated mTOR activity and increased levels of ferritin H (ferritin H is mainly removed from cells by the autophagy–lysosome pathway)32. In addition, emerging evidence in aged rats has highlighted an age-associated decline in expression of the autophagy-related protein beclin 1 (BECN1) in whole brain tissue, as well as in the hippocampus of naked mole rats and Wistar rats33,34. In line with observations in rodent models, findings in humans have suggested that the expression of autophagy-related genes, such as ATG5, ATG7 and BECN1, declines with age35. Moreover, the development and progression of several human pathologies is highly associated with age-dependent autophagy deficits19,36,37. Collectively, these studies demonstrate that a gradual decline in the abundance of autophagy-related proteins and reduced delivery of cargo to lysosomes occur with age, implicating compromised autophagy as a cardinal feature of organismal aging.
In line with a causal role for autophagy in the aging process14, genetically impairing nonselective or selective autophagy results in accelerated tissue functional decline and disease in a range of experimental models. Transcriptomic profiling in Saccharomyces cerevisiae has provided evidence of defective autophagy among short-lived as compared to long-lived mutants38. In addition, selective mutation(s) and/or knockdown of genes encoding components of the autophagic machinery in C. elegans (lgg-1 (ortholog of ATG8), unc-51 (ortholog of ATG1), bec-1, atg-7, lgg-3 (also known as atg-12) and atg-18), Drosophila (Atg3 and Atg8a) and mice (Atg5, Atg7 and Becn1) shorten lifespan and healthspan1,14,30,39. In line with these observations, systemic genetic knockout of autophagy components (Becn1, Atg5, Atg9 and Atg13) is lethal in mice, highlighting the importance of autophagy in development40. Furthermore, knockdown of genes encoding transcription factors that regulate autophagy, such as TFEB (ortholog in C. elegans, hlh-30) and FOXO (encoding forkhead box O; ortholog in C. elegans, daf-16) shortened lifespan in both wild-type worms and long-lived daf-2 (insulin/insulin-like growth factor-1 (IGF-1) receptor) mutants41.
In contrast, studies in long-lived mutant animals have shown that increased autophagy is associated with delayed aging. In particular, the extended lifespan of C. elegans daf-2 loss-of-function mutants is dependent on autophagic genes, such as bec-1, lgg-1, atg-7 and atg-12 (refs. 1,14,42). Furthermore, HLH-30 is required for the long lifespan of multiple longevity mutants, including not only daf-2 mutants with reduced insulin/insulin-like signaling, but also germline-less glp-1(e2141) mutants, dietary-restricted eat-2(ad1116) mutants, mitochondrial respiration-defective clk-1(e2519) mutants and mRNA translation-impaired rsks-1(sv31) mutants43. These findings coincide with impaired induction of autophagosome formation and lysosomal degradation upon loss of hlh-30, suggesting that HLH-30 promotes longevity by regulating the autophagy process downstream of multiple lifespan extension paradigms43. In addition, formation of long-lived dauer worms, corresponding to a larval hibernation stage, is also associated with increased autophagy and is dependent on the autophagy genes atg-1, atg-7, lgg-1 and atg-18, underlining the essential role of autophagy in organismal adaptation during challenging conditions42.
In line with observations from long-lived mutants, genetic or pharmacological upregulation of autophagy promotes longevity in animals. Autophagy induction by overexpression of Atg genes in Drosophila (Atg1 and Atg8a) and mice (Atg5) extends lifespan30,44,45. Similarly, Bcl2 mutations that disrupt the BECN1–BCL-2 complex increase basal autophagic flux, which results in long-lived male and female mice with improved healthspan46. Overexpression of autophagic regulators in C. elegans and Drosophila, such as AMPK, further facilitates autophagy in diverse tissues and in turn extends longevity14,45. Additionally, hlh-30 overexpression enhances autophagy and promotes lifespan extension in C. elegans43, and silencing of the nuclear export protein exportin-1 (XPO-1) enhances autophagy by enrichment of HLH-30 in the nucleus, which is accompanied by proteostatic benefits and improved longevity47. Moreover, rapamycin, an inhibitor of the mTOR pathway, has been shown to extend the median and maximum lifespan of both female and male mice when fed to them late in life48.
Accumulating evidence in aged mice, as well as in rodent models recapitulating characteristic features of human diseases, has shown that compromised autophagy is among the most common factors contributing to the collapse of tissue homeostasis. In particular, age-associated dysregulation of autophagy (demonstrated by the accumulation of autophagosomes), possibly due to impaired lysosomal fusion and/or degradation, is associated with cellular dysfunction and/or death, which contribute to neurodegeneration, as well as cardiac and skeletal muscle aging49,50,51,52,53. In hematopoietic stem cells (HSCs), autophagy has been shown to delay aging via activation of downstream sirtuin-3 (SIRT3), a key mitochondrial protein capable of rejuvenating blood and protecting against oxidative stress in mice and human HSC-enriched cells54.
Moreover, autophagy appears to be a critical mechanism to maintain immune memory in mice, and levels of the endogenous autophagy-inducing metabolite spermidine fall in human T cells with age. In fact, supplementation of T cells from older donors with spermidine restores autophagy levels to those observed in younger donors via the eIF5A translation factor and TFEB transcription factor55. Furthermore, spermidine administration in a mouse model of mild cognitive impairment, a transitional phase between healthy aging and AD, led to an improvement in degradation of misfolded proteins and an accompanying delay in age-related memory deficits, thereby implicating autophagy as a pathophysiological mechanism of action56.
While dysregulation of autophagy underlies aging and disease phenotypes, excessive autophagy may also contribute to the deterioration of cellular function in some contexts. Recent evidence has demonstrated that an age-dependent decline in the levels of Rubicon, a negative regulator of autophagy, exacerbates metabolic disorders in adipocytes57. While strongly upregulated autophagy may exacerbate metabolic disorders, this finding may also be attributed to autophagy-independent changes in metabolism. Furthermore, elevated autophagy has been found to shorten lifespan in C. elegans mutants lacking sgk-1 (encoding serum/glucocorticoid-regulated kinase-1). Loss of this kinase results in increased mitochondrial permeability, leading to excessive autophagy and reduced organismal fitness in worms and mice58. Conversely, reducing the levels of autophagy in sgk-1 mutants or suppressing the opening of the mitochondrial permeability transition pore restores normal lifespan58. Similarly, suppressing autophagy exclusively in the intestine of post-reproductive adults at higher temperatures has been proposed to prevent the emergence of age-related pathologies in C. elegans59. However, it should be noted that this is in direct contrast to findings in long-lived mutants, where intestinal autophagy is enhanced60,61. Another study in C. elegans showed that short interfering RNA (siRNA)-based reduction in the abundance of the VPS-34–BEC-1–EPG-8 autophagic nucleation complex in aged post-reproductive worms extended lifespan and improved neuronal integrity29. However, detailed data on knockdown efficiency in aged worms, as well as an understanding of the remaining levels of neuronal autophagy, are necessary to ensure accurate in-depth data interpretation. Collectively, these observations suggest that maintenance of functional autophagy is essential for healthy cellular and organismal aging and that dysregulation of autophagy in either direction, whether insufficient or excessive, contributes to cellular deficits and functional organismal decline.
A summary of autophagy-related genes linked to longevity and disease is provided in Table 1 and Supplementary Table 1. In addition, several interventions known to promote lifespan, including dietary restriction and treatment with pharmacological agents, such as rapamycin, spermidine and NAD+ precursors, require intact autophagic machinery. In totality, these findings reinforce the notion that autophagy stimulation is necessary and sufficient to sustain organismal homeostasis and extend longevity in multiple model organisms (discussed in detail below)1. An overview of autophagy inducers linked to enhanced longevity and improved health is presented in Table 2.
Together, numerous studies have provided evidence that (1) autophagy is compromised during the process of aging; (2) dysfunction of autophagy shortens lifespan in various experimental animal models; and (3) promotion or restoration of autophagy contributes to lifespan and healthspan extension in diverse organisms. This suggests that autophagy is a central regulator of aging. However, an important and fundamental question remains unanswered: how does autophagy facilitate long-term cell and tissue health?
The multifaceted role of autophagy in health and aging
Autophagy and protein homeostasis
Protein homeostasis (proteostasis) collapse is a central hallmark of aging and disease that is characterized by the appearance of misfolded, mislocalized and aggregated proteins. While age-related loss of proteostasis has been documented in numerous tissues, age-dependent protein aggregation is strongly linked to neurodegenerative pathologies such as AD, Parkinson’s disease, Huntington’s disease and amyotrophic lateral sclerosis (ALS)62,63.
Along with molecular chaperones and the ubiquitin–proteasome system (UPS), autophagy is a central regulator of cellular proteostasis that operates to (1) degrade soluble misfolded or oligomeric proteins via CMA (the selective degradation of ubiquitin-tagged protein aggregates by chaperone-assisted selective autophagy) and (2) remove bulk protein aggregates13,19,64. In line with this, genetic perturbation of core components or regulators of the autophagy machinery accelerates age-related protein aggregation, shortens lifespan and exacerbates pathological features in worm, fly and mouse models of disease. Conversely, increasing autophagy, genetically or pharmacologically, suppresses protein aggregation and promotes health and longevity48,65,66 (also reviewed in refs. 67,68,69).
In C. elegans, loss-of-function mutations in bec-1 or atg-18 or RNA interference (RNAi) against bec-1, atg-9 or lgg-1 increases susceptibility to protein aggregation, accelerates the onset of age-related paralysis and shortens lifespan39,70. Similarly, mutations in the core autophagy components Atg8a or Atg7 in Drosophila increase the levels of insoluble protein aggregates and reduce longevity30,71. Finally, knockout of Atg5 or Atg7 in mouse neurons leads to the appearance of cytoplasmic inclusion bodies in the brain and early-onset neurodegeneration72,73,74, while knockout of Lamp2 (the primary receptor for CMA) in the liver results in altered proteostasis and hepatic dysfunction with age75.
Conversely, enhanced proteostasis and extended lifespan in C. elegans occur when the lysosome–autophagy transcription factor HLH-30 or the selective autophagy receptor p62/SQSTM-1 is upregulated. Likewise, in Drosophila, overexpression of Ref(2)P (p62 ortholog) or the autophagy activator FOXO reduces protein aggregation in various tissues and extends lifespan43,76,77,78,79. Pharmacological (for example, via clonidine, rilmenidine or rapamycin) or genetic (for example, atg5) upregulation of autophagy in zebrafish harboring the rare p.Ala152Thr variant of tau ameliorates tau pathology80. Increased autophagy is also associated with enhanced clearance of protein aggregates in mammals, as systemic overexpression of Atg5 or Becn1 genes with mutations that disrupt BECN1–BCL-2 binding improves proteostasis and promotes longevity in mice44,81, while overexpression of the selective autophagy mediator BAG3 suppresses tau accumulation in neurons82.
As a complement to the genetic modulation of autophagy, treatment of Drosophila with the mTOR inhibitor rapamycin suppresses age-related protein aggregation and extends lifespan in an autophagy-dependent manner83. Furthermore, in cell culture and fly models, rapamycin suppresses toxicity from neurodegenerative disease-associated proteins, including mutant huntingtin, polyalanine-expansion-containing proteins and tau84. Several other pharmacological autophagy inducers, such as spermidine and nicotinamide, have also been reported to protect against proteostasis collapse and proteotoxicity in various models of Huntington’s disease, AD, Parkinson’s disease and ALS67.
Autophagy has also been linked to stem cell function, with the autophagy-mediated clearance of protein aggregates central to the activation of quiescent neuronal stem cells. Activating autophagy by overexpression of TFEB or rapamycin supplementation inhibits age-related protein aggregation and enhances neuronal and muscle stem cell function in aged mice85,86,87. Given the fact that stem cell exhaustion is intimately linked to age-related tissue dysfunction, these findings suggest that enhancing proteostasis specifically in stem cells may preserve many aspects of healthy tissue function during aging. Collectively, these observations strongly support the notion that autophagy promotes healthy aging by protecting cells against toxic misfolded and aggregated proteins.
Autophagy regulation of macromolecule availability
Another important role for autophagy in cellular homeostasis and organismal aging is to ensure the availability of metabolites, including amino acids, lipids, carbohydrates and nucleic acids, especially during states of stress, such as nutrient starvation (Fig. 1a). Under challenging conditions, autophagy promotes cellular metabolism and survival by recycling amino acids, which are generated from the degradation of cytosolic substrates, to replenish nutrients, produce energy and promote protein synthesis. An inability to properly recycle amino acids through autophagy is linked to growth and developmental defects in Atg5-deficient mice and impaired growth during nitrogen starvation in several atg-deficient yeast cell lines (including atg1Δ, atg2Δ, atg7Δ, atg11Δ, atg15Δ and atg32Δ mutants) 88,89,90. Autophagy can also be tailored to mediate the availability of carbohydrates, lipids and nucleic acids through three main cellular processes: glycophagy, lipophagy and RNAutophagy or DNAutophagy, respectively.
Glycophagy
Glucose is the primary energy source for cellular metabolism. It is stored as glycogen, and metabolism of glucose is tightly regulated in a tissue-dependent manner (that is, the liver maintains blood glucose levels, while muscles are the source of cellular energy). However, various conditions resulting in metabolic stress, such as starvation, stimulate glycogen breakdown to augment cellular glucose levels and promote metabolic activity91. Glycogen can be degraded in the cytosol through the activity of glycogen phosphorylase and glycogen debranching enzymes (detailed in ref. 92) or in the lysosome via autophagy. The selective clearance of glycogen via autophagy, referred to as glycophagy, has a crucial role in glucose homeostasis. In response to nutrient deficiency, the energy sensor AMPK is activated, which in turn inhibits mTOR complex 1 (mTORC1), leading to activation of the ULK1 kinase, which is important for induction of autophagy (AMPK–mTORC1–ULK1 triad)91. Recent findings in yeast have demonstrated that Atg11 is necessary to facilitate interaction between the AMPK homolog Snf1 and the ULK1 homolog Atg1 upon glucose starvation to promote autophagy93. The LC3-interacting region (LIR) motif, also known in yeast as the Atg8-family-interacting motif (AIM), in starch-binding domain-containing protein 1 (STBD1) may allow cells to physically link glycogen to GABARAPL1, facilitating the transport of glycogen to lysosomes for degradation (Fig. 1a, pathway (3))94. In parallel to glycophagy, other pathways such as β-oxidation may maintain cellular bioenergetics to compensate for glucose deprivation95. Autophagy also has a pivotal role in maintaining cell function, not only in glucose starvation but also in conditions of excess glucose. High glucose levels were associated with mitochondrial dysfunction, generation of reactive oxygen species and induction of autophagy in endothelial progenitor cells96.
Under conditions of impaired autophagy, accumulation of glycogen contributes to the pathogenesis of age-related diseases. In Pompe disease, a lysosomal storage disorder, the ability of lysosomes to degrade glycogen is impaired, owing to a deficiency in the lysosomal hydrolytic enzyme acid α-glucosidase (GAA). This results in accumulation of lysosomal glycogen in many tissues, predominantly in skeletal and cardiac muscle, leading to progressive lethal skeletal myopathy and respiratory and cardiac defects97. Impaired tissue function from the inability of lysosomes to degrade glycogen also leads to energy deficiency in skeletal muscle. For infantile-onset Pompe disease98, a promising therapeutic intervention is administration of recombinant human GAA. Furthermore, dysfunctional autophagy-mediated accumulation of glycogen has been demonstrated to be the cause of neurodegeneration in a mouse model of Lafora disease, with this accumulation suppressed when glycogen synthase is deleted99. These findings indicate that glycogen accumulation might be a cause, rather than a consequence, of impaired autophagy, resulting in impaired cellular function and disease. Glycophagy, therefore, is essential for cellular function and survival, suggesting that levels of glycophagy could determine organismal health and possibly longevity.
Lipophagy
Intracellular storage and use of lipids is critical to maintain cellular energy homeostasis. In response to starvation, triglycerides stored in lipid droplets are hydrolyzed by specific lipases into free fatty acids for energy metabolism. Lipid droplets can also undergo selective degradation by autophagy, termed lipophagy, as an alternative mechanism for regulating lipid homeostasis (Fig. 1a, pathway (4))100. Thus far, a specific receptor coupling lipid droplets to autophagosomes and trafficking to lysosomes has not yet been identified, although LC3-mediated engulfment of lipid droplets has been observed101. Moreover, CMA has been implicated in degradation of the lipid droplet-associated proteins perilipin 2 (PLIN2) and perilipin 3 (PLIN3)102. Lysosomal acid lipases are involved in the degradation of lipid droplets; in particular, lipolysis is conducted primarily by adipose triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL), and selective knockdown of ATGL and HSL in mice results in selective inhibition of lipid droplet degradation, while other autophagy processes (that is, degradation of proteins and organelles) serve as a compensatory mechanism to replenish the reduced availability of energy substrates103. An age-dependent decline in basal autophagy in the liver may underlie the accumulation of hepatic lipids, which in turn has been proposed to contribute to metabolic conditions as well as impairing autophagy, a vicious cycle promoting aging100. For example, age-dependent reduction in CMA is likely due to alterations in the lipid composition of discrete microdomains at the lysosomal membrane, including altered dynamics and stability of the CMA receptor LAMP2A in the lysosome104. Additional mechanisms by which age-related alterations in lipid composition and/or levels may impair autophagy remain unknown. Further, age-dependent accrual of lipid droplets and ectopic fat deposition are highly interconnected with the age-dependent decline in autophagy and/or autophagic defects105,106. Autophagy and LIPL-4-dependent lipolysis are both upregulated in germline-less C. elegans and work interdependently to prolong lifespan107. The mammalian homolog of worm LIPL-4 is lysosomal acid lipase (LIPA), a key enzyme involved in the hydrolysis of cholesterol via autophagy108,109. Cellular supplementation with NAD+, which stimulates autophagy and subtypes of autophagy, including mitophagy, and stimulates the activity of the NAD+-dependent sirtuin-1 (SIRT1) and SIRT3 pathways, reduced fat accumulation and increased lifespan in progeroid animals fed a high-fat diet110,111,112, highlighting the importance of autophagic degradation of lipids in healthspan and lifespan.
In pathological conditions such as alcoholic fatty liver disease (AFLD), impaired lipophagy has been shown to be the basis of lipid peroxidation and cellular damage. AFLD results from excessive consumption of alcohol, leading to damage to the liver in the form of oxidative stress, excessive lipid droplet accumulation in the cytoplasm of hepatocytes (steatosis), mitochondrial damage and cell death. Acute exposure to ethanol triggers lipophagy, which acts as a defense mechanism against lipid peroxidation, thereby protecting hepatocytes. However, chronic exposure to ethanol leads to mTOR-mediated inhibition of lipophagy, which in turn contributes to lipid peroxidation and cell death22,113,114. In fact, inhibition of mTOR-mediated suppression of TFEB, using torin-1, resulted in enrichment of TFEB levels in the liver and protection against steatosis and ethanol-induced liver injury115. Genetic overexpression of TFEB in the liver was shown to increase lysosomal biogenesis and enhance mitochondrial bioenergetics, which served as a protective mechanism against ethanol-induced liver injury in mice. In line with these findings, knockdown of TFEB in the liver of mice resulted in more severe liver injury in response to increased ethanol consumption115. In addition, lipophagy is key for the differentiation of several cell types, including hepatocytes116 and neutrophils117. Knocking out ATG7 in HSCs leads to an accumulation of immature neutrophils resembling the myeloid bias of an aging hematopoietic system. Differentiation can be rescued by supplementation with exogenous free fatty acids used for β-oxidation, further demonstrating that lipophagy usually provides these during the energy-intensive process of differentiation. Further studies on the molecular mechanisms of lipophagy, including identification of lipid-specific autophagy receptors and their impact on cellular homeostasis, will shed light on the relationship among autophagy, metabolism and aging.
Autophagic degradation of nucleic acids: RNAutophagy and DNAutophagy
Nucleic acids are degraded via multiple mechanisms (a complete description of which is beyond the scope of this Review; see details in refs. 118,119), including by autophagy. RNA and DNA are targeted for lysosomal degradation via several pathways, including LC3-dependent autophagic degradation of stress granules (condensates of protein and RNA)120, p62- and NDP52-dependent autophagic degradation of retrotransposon RNA121, lysosomal membrane protein LAMP2C-dependent direct binding to RNA (also DNA122) for lysosomal degradation123 and a lysosomal putative RNA/DNA transporter, SID1 transmembrane family, member 2 (SIDT2), that mediates direct uptake of RNA (and DNA124) for lysosomal degradation125. At present, little is known about whether or how RNAutophagy (also known as RNAphagy) and DNAutophagy (also known as DNAphagy) affect health and aging. However, it is reasonable to suggest that nucleic acid turnover is essential for health, as accumulation of damaged or unnecessary DNA and RNA in the cytosol promotes inflammation, cancer and even accelerated aging68,126,127. DNA damage triggers autophagy and subtypes of autophagy that are considered to be cell survival responses128; in contrast, genetic or age-dependent impairment of DNA repair leads to genomic instability, cellular dysfunction, cell death and accelerated aging68. Exogenous DNA or RNA (for example, microbial) or endogenous nuclear or mitochondrial DNA in the cytoplasm may trigger autophagy. Nuclear DNA (including extranuclear chromatin) could be aberrantly released into the cytoplasm as a result of impaired nuclear envelope integrity, nuclear envelope blebbing or nuclear export processes129, while mitochondrial DNA could leak into the cytoplasm as a result of mitochondrial damage and inefficient elimination of damaged mitochondria via mitophagy126,127. The cyclic GMP-AMP (cGAS)–stimulator of interferon genes (STING), or RIG-I–MAVS, signaling axis detects these nucleic acid fragments to initiate an innate immune reaction, linking it to autoimmunity, inflammation, senescence and autophagy129. Collectively, genomic instability, accumulation of mitochondrial DNA leakage in the cytoplasm and increased levels of cellular stress granules are linked to inflammation, accelerated aging and a broad range of neurodegenerative diseases120,121,126. Although maintenance of DNA and RNA homeostasis is critical for healthy aging, the contribution of RNAutophagy and DNAutophagy to long-term tissue health and pathology requires further exploration.
Autophagy of subcellular organelles: mitophagy, ER-phagy, nucleophagy and lysophagy
Aging is associated with an accumulation of damage to subcellular organelles. Timely and efficient disposal and recycling of dysfunctional organelles is necessary to maintain cellular function and viability. Selective autophagy is the common mechanism underlying the clearance of damaged and/or superfluous subcellular organelles such as mitochondria (mitophagy), the ER (reticulophagy or ER-phagy), the nucleus (nucleophagy) and lysosomes (lysophagy)17. Both membrane-bound and soluble selective autophagy receptors are involved in the selective degradation of organelles18,130.
Among the different types of autophagy targeting subcellular organelles, the most investigated is mitophagy. Mitophagy is the selective autophagic elimination of defective or surplus mitochondria. The PINK1- and parkin-mediated pathway for degradation of heavily depolarized mitochondria is best understood and involves attraction by Ser65-phosphorylated ubiquitin of the soluble selective autophagy receptors NDP52, optineurin and p62, which then recruit the core autophagy machinery for autophagosome formation on the damaged mitochondria131. Other basal, developmental and stress-induced mitophagy pathways involve binding of LC3 to a series of LIR-containing mitochondrial outer membrane proteins, such as NIX (BNIP3L), BNIP3, FKBP8, FUNDC1, BCL2L13, PHB2 and AMBRA1, as well as LC3-binding mitochondrial lipids such as cardiolipin37 (Fig. 1a, pathway (6), left). While whole mitochondria can be degraded via mitophagy, it appears that organelles with minor damage can be ‘repaired’ by other quality-control mechanisms such as the piecemeal mitophagy pathway, which is a basal housekeeping mitophagy pathway that involves degradation of mitochondrial proteins in an LC3C- and p62-dependent manner132 (Fig. 1a, pathway (6), right). Other mitochondrial degradation pathways include the mitochondria-derived vesicle (MDV) pathway, where damaged cargo (for example, impaired mitochondrial proteins) is delivered to the lysosome for degradation in a process dependent on syntaxin-17, PINK1 and parkin133. A recent study in C. elegans showed that damaged subcellular components, including mitochondria, can be budded off from certain neurons via membrane-bound vesicles (termed ‘exophers’)134. Once in the extracellular space, these damaged organelles can be engulfed and digested by surrounding cells134. This cellular release of exophers is conserved in mammals, as cardiomyocytes release exophers (containing mitochondria) to be received and eliminated by adjacent macrophages135.
Accumulating evidence has highlighted that mitophagy is a critical contributor to cellular physiology and organ homeostasis. First, there is an increase in mitophagy from juvenile stages to adulthood, followed by a dramatic reduction in aged animals. For example, there is an increase in basal mitophagy levels in fly flight muscles from the ages of 1 week to 4 weeks136; in mice, mitophagy in the dentate gyrus (DG), a region of the brain that is essential for memory, was reduced by approximately 70% between the ages of 3 and 21 months137. Mitophagy is also impaired under high-fat feeding conditions137 and in neurodegenerative diseases (reviewed in ref. 37). Indeed, mitophagy is reduced in mice with AD (by approximately 50% in the hippocampus in comparison to healthy controls)138, Parkinson’s disease (reviewed in ref. 139) and Huntington’s disease (by over 70% in the DG of huntingtin-expressing mice versus wild-type controls)137. Second, intact mitophagic machinery is required for longevity. Because there are several redundant mitophagy pathways, dysfunction of isolated individual mitophagy pathways may not affect lifespan140,141. However, mitophagy is essential for longevity under conditions of low insulin/IGF-1 signaling (C. elegans daf-2 mutants) and dietary restriction (C. elegans eat-2 mutants)140,142, as well as for the maintenance of neuronal functions in response to stressful conditions126. Third, mitophagy induction is sufficient to improve healthspan and extends lifespan in several model organisms, rescues age-associated neurodegenerative phenotypes in AD138,143 and prolongs lifespan in nematode and fly models of accelerated aging66,111,144. Moreover, functional mitophagy is essential for restraining innate immunity, as mitochondrial stress can lead to the release of damage-associated molecular patterns (DAMPs) that activate innate immunity. Inflammation resulting from excessive exercise in Pink1- and Parkin-knockout mice has been shown to be suppressed by loss of STING, a central regulator of the type I interferon response to cytosolic DNA126.
Other autophagic pathways that target subcellular organelles include ER-phagy, nucleophagy and lysophagy. In yeast, Atg39 regulates perinuclear ER-phagy and nucleophagy, while Atg40 is necessary for cortical and cytoplasmic ER-phagy145 (Fig. 1a, pathway (7)). ER-phagy is conserved in mammalian cells through specific ER-phagy receptors, such as FAM134B, SEC62, RTN3L, CCPG1, ATL3 and TEX264 (reviewed in ref. 146). Nucleophagy is conserved in mammalian cells147 and involves nuclear LC3B–lamin B1 interaction-based nuclear-to-cytoplasmic degradation, which may be a defense mechanism protecting cells from tumorigenesis148 (Fig. 1a, pathway (8)). Lysophagy is regulated by both ubiquitin-dependent (galectin-3–TRIM16–ULK1–autophagy receptor–LC3, the F-box protein FBXO27 and UBE2QL1) and ubiquitin-independent (galectin-8–autophagy receptor–LC3) pathways (reviewed in ref. 149) (Fig. 1a, pathway (11)). Maintenance of functional and effective lysosomes, via timely and efficient lysophagy, is essential for cell survival. In particular, dysfunction in lysosomal membrane proteins such as SCAV-3, the C. elegans homolog of human LIMP-2, has been linked to reduced lifespan, implicating lysosome integrity as a defining factor in longevity150,151,25. Moreover, dysfunctional lysosomal membrane proteins coupled to leakage of proteolytic enzymes (that is, cathepsin D) into the cytosol have been associated with aging and pathological aging in a broad range of neurodegenerative diseases152. Thus, maintaining physiological lysophagy is critical for many cellular processes and is presumably important for health and longevity, as lysosomal rupture triggers endolysosomal damage responses and even lysosomal cell death, which is linked to aging and diseases152,153.
Collectively, an imbalanced quality surveillance system for subcellular organelles, such as mitochondria, the ER, small nuclear fractions and lysosomes, might be a causative factor for age-related pathologies as well as premature aging. Further studies on mitophagy, ER-phagy, nucleophagy and lysophagy to decipher their multilayer regulatory network and association with aging and health are necessary. In particular, studies to address how these processes change with age and how they influence age-related tissue function will lead to critical insights with broad relevance to human health and quality of life.
Xenophagy
Xenophagy (from the Greek meaning ‘to eat foreign matter’) is the process by which autophagy targets pathogens154. Many pathogens are known to be degraded by autophagy, while others take over core autophagy components for their own benefit155 (Fig. 1a, pathway (9)). Indeed, several studies have demonstrated that autophagy can target bacteria such as Rickettsia conorii156, Listeria monocytogenes157, Streptococcus pyogenes158 and Mycobacterium tuberculosis159,160. Xenophagy may also protect the body against invasion by viruses and parasites.
Upon their intake by inhalation, M. tuberculosis bacteria are captured by alveolar macrophages. However, they have evolved the ability to impair phagosome maturation (which under normal conditions would lead to phagocytosis) and end up hijacking macrophages161. Later on, using secretions from ESX-1 (6-kDa early secretory antigenic target (ESAT-6) secretion system 1), the bacteria are able to break free from the phagosome and enter the cytosol. Here xenophagy comes into action. cGAS detects the bacterial DNA162, which results in ubiquitination of the invading bacteria by Smurf1 (or parkin)163. NBR1 (or p62) attaches to these ubiquitin chains, resulting in the recruitment of LC3B and, ultimately, autophagic degradation164. Indeed, an absence of autophagic machinery components, in particular, ULK1 (ref. 165), BECN1 (ref. 166), p62 (ref. 167), ATG7 (ref. 168) and TBK1 (ref. 167), may promote proliferation of the bacteria. The mechanism is similar for specific viruses. BECN1 and p62 in selective autophagy of viral capsids can be protective against Sindbis virus169,170. However, other viruses, such as herpes simplex virus type 1, have evolved to inhibit autophagy by targeting BECN1 (ref. 171). In addition, several studies have highlighted the importance and possible therapeutic relevance of autophagy for controlling severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (COVID-19)172,173,174. With respect to parasites, autophagy can control Toxoplasma gondii, while knockout of ATG5, ATG7 or ATG16L1 renders mice more likely to succumb to parasites175. A detailed review of the relationship between parasites and autophagy is available176.
Although there are not many data available on a direct link between xenophagy and aging or lifespan, it is conceivable that blocking infection by exogenous intruders is required for maintenance of a healthy state and reduced inflammation151,177. Further work to investigate the molecular mechanisms of xenophagy and their association with aging and longevity is required.
Tissue-specific autophagy in aging
As aging is associated with functional decline at both the tissue and organismal level, it is important to understand how aging within individual tissues affects, and is affected by, aging across the entire organism. Evidence from nematodes, flies and mice has revealed that autophagy may have tissue-specific roles in regulating aging14. Inhibition of lgg-1 and atg-18 specifically in the body wall muscle of adult worms is sufficient to shorten the lifespan of the long-lived dietary-restricted eat-2 and insulin/IGF-1 receptor-deficient daf-2 mutants28,178. In addition, the shortened lifespan in atg-18 mutants (ATG-18 is a member of the WIPI protein family, homologous to mammalian WIPI-1 and WIPI-2) can be suppressed by tissue-specific restoration of ATG-18 function: pan-neuronal or intestine-specific expression of atg-18 fully restored the lifespan of atg-18 mutants to that of wild-type worms, while muscle- or hypodermis-specific rescue of ATG-18 had little to no ability to restore lifespan179. In flies, promotion of autophagy in muscle tissue via overexpression of Atg8a or the transcription factor FOXO was sufficient to extend lifespan78,180, while, in mice, inhibition of autophagy through muscle-specific ATG7 deficiency resulted in impaired muscle function (possibly via mitochondrial dysfunction) and decreased lifespan181. Furthermore, enhancing autophagy specifically in the intestine results in maintenance of intestinal barrier function and promotes longevity and healthspan in worms and flies45,178. Given that tissues age unevenly, with some tissues presenting with faster degeneration than others182, it will be interesting to determine how closely rates of aging and autophagy are correlated in different tissues throughout life.
Defective autophagy in diseases associated with accelerated aging, neurodegenerative diseases and inflammaging
Accumulating evidence from studies using laboratory animals and human samples supports an essential role for autophagy in embryonic development, tissue health and lifespan through the suppression of age-associated inflammation (inflammaging), maintenance of genomic integrity, preservation of cellular and tissue homeostasis, and rejuvenation of stem cells (Fig. 3a; refs. 13,14,68,69). While autophagy is tightly regulated by multiple molecular pathways involving central modulators of energy metabolism, such as AMPK, mTORC1, sirtuins and calcineurin (Fig. 3b), several interventions such as dietary restriction, exercise and supplementation with small chemical compounds (detailed below) stimulate autophagy69. Recent preclinical studies have linked impairment of general autophagy or subtypes of autophagy (in some diseases, while a subtype of selective autophagy is impaired, there may be no change, or even an increase, in general autophagy) to pathological states such as progeria and a series of accelerated aging diseases68 (Fig. 3c), neurodegenerative diseases19,37 (Fig. 3d) and other disorders13,14,68,69. For example, maintenance of CMA in aged cells sustains HSC function183 and prevents collapse of the neuronal metastable proteome184. Similarly, mitophagy, which is reduced in both normal aging and AD, extends healthspan140 and suppresses amyloid β- and phosphorylated tau-induced memory loss when stimulated in aged tissues138.
a, Autophagy participates in multiple processes that are essential for longevity. b, A brief summary of some of the major known mechanisms that regulate autophagy in multiple organisms and their influence on the process. c, A list summarizing premature aging diseases with impaired mitophagy as a cause of mitochondrial dysfunction, which contributes to short lifespan (LS) and healthspan (HS). These premature aging diseases are ataxia telangiectasia (AT), Cockayne syndrome (CS), Fanconi anemia (FA), Hutchinson–Gilford syndrome (HG), Werner syndrome (WS) and xeroderma pigmentosum (XP; especially group A). Changes in autophagy and mitophagy in Hutchinson–Gilford syndrome are elusive. d, Autophagy (including subtypes of selective autophagy, such as mitophagy) is impaired in broad neurodegenerative diseases, where impairment may drive or exacerbate disease progression. These diseases include AD, Parkinson’s disease (PD), Huntington’s disease, ALS and frontotemporal dementia (FTD). We emphasize that these are not the only drivers of the diseases and other processes may have roles leading to pathology and symptomatology.
Understanding the relationship between compromised autophagy and other hallmarks of aging will provide a better understanding of the molecular events that promote aging and disease14,68,69. Among the many age-related changes previously described, inflammation is linked to autophagy, as impaired autophagy results in inflammation, and has emerged as a major driver of age-related tissue damage63,69,185,186. Inflammation is an evolutionarily conserved protective mechanism designed to maintain organismal homeostasis in the face of acute and local perturbations and serves as an adaptive response to infection or injury187. Chronic, systemic inflammation develops progressively with age and contributes to organismal deterioration through the process termed inflammaging186.
Autophagy has been identified as one of the pivotal mechanisms orchestrating the differentiation and metabolic state of innate immune cells. In particular, the balance between mTOR and AMPK activation has a central role in immune cell maintenance and function. Upon mTOR activation, autophagic flux is reduced, accompanied by increased cellular glycolytic activity, giving rise to a proliferative and pro-inflammatory phenotype in macrophages. In contrast, AMPK activation drives autophagy and promotes the OXPHOS-dependent function of non- or anti-inflammatory macrophages188.
Autophagy also regulates the NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome, which is an intracellular protein complex that activates caspase-1, which in turn catalyzes the cleavage, activation and subsequent release of pro-inflammatory cytokines (for example, interleukin (IL)-1β), which can induce neurodegeneration186,189. The NLRP3 inflammasome has been identified as a critical component of the innate immune response (that is, the response to microbial motifs, endogenous danger signals and environmental irritants) and orchestrates host immune homeostasis189. Defective autophagy, for example, in models of selective knockout or knockdown of genes encoding components of the autophagic core machinery (for example, ATG5, ATG7, BECN1 and MAP1LC3B), results in unrestricted inflammasome activation and consequent inflammation. Likewise, promotion of autophagy through starvation or with pharmacological agents (for example, rapamycin) inhibits the inflammasome190. In addition, evidence stemming from an APP/PS1 mouse model of AD demonstrated mitophagy-induced inhibition of the NLRP3 inflammasome, resulting in reduced neuroinflammation138. These findings imply an important role for autophagy in the regulation of inflammation and, in turn, aging and neurodegenerative diseases.
Anti-aging effects of autophagy modulators
The mounting evidence that an imbalance of autophagy is an important age-associated characteristic has driven extensive research into the development of compounds that can promote autophagy1. Pharmacological agents promoting autophagy can be classified on the basis of their effect on the mTOR pathway191. mTOR inhibition by rapamycin has been shown to reduce protein synthesis and promote autophagy, both of which contribute to extended lifespan in yeast, nematodes, flies and mice (Table 2). In addition, rapamycin has been demonstrated to protect against neurodegenerative diseases, including AD, via promotion of autophagy; however, rapamycin treatment was observed to be detrimental in the case of models of ALS, possibly owing to non-autophagy-related side effects191. Other pharmacological agents reported to promote autophagy via direct interaction with mTOR include torin-1 and PP242 (ref. 192). mTOR-independent promoters of autophagy mainly act via the AMPK pathway. Examples include metformin and trehalose, which have been demonstrated to be effective in enhancing autophagy, extending lifespan and protecting against neurodegeneration in experimental models191.
Compounds such as resveratrol and spermidine modulate the acetylation state of proteins to regulate autophagy and promote longevity. Resveratrol is a natural polyphenol that reportedly promotes lifespan in C. elegans and healthspan in mice via activation of the NAD+-dependent deacetylase SIRT1 (refs. 112,193,194). Spermidine is a polyamine that extends the lifespan of yeast, worms, flies and mice by enhancing autophagy through inhibition of the EP300 acetyltransferase195, among other mechanisms55,196,197,198. The longevity-extending effects of spermidine are abolished upon depletion or deletion of essential autophagy genes such as bec-1 in C. elegans and Atg7 in yeast and flies197,199. Furthermore, pharmacological inhibition of XPO-1 results in enhanced autophagy (as evidenced by an increase in the frequency of autophagosomes and autolysosomes) and increased lifespan in C. elegans. These effects were mediated by nuclear enrichment of HLH-30, which occurred in an mTOR-independent manner47. Additional modulators of TFEB homologs that regulate autophagy and have also been demonstrated to protect against pathophysiological aging include ouabain and fisetin. Ouabain is a cardiac glycoside that enhances activation of TFEB through inhibition of the mTOR pathway and induces downstream autophagy–lysosomal gene expression and cellular restorative properties200. Ouabain has been shown to reduce the accumulation of abnormal toxic tau both in vitro and in vivo200. Fisetin is a flavonol and was shown to facilitate the clearance of endogenous tau via TFEB (through inhibition of mTOR kinases) and Nrf2 activation20.
Other small molecules that induce subtypes of autophagy, especially mitophagy, also enhance longevity and suppress age-associated diseases. These include NAD+, a fundamental metabolite in energy metabolism, redox homeostasis, mitochondrial function, and the arbitration of cell survival and death185. NAD+-activated sirtuins stimulate autophagy via mTOR inhibition and deacetylation of several key autophagy proteins (ATG5, ATG7 and ATG8)201,202. In addition, the NAD+–SIRT axis activates mitophagy by increasing the activity of a series of mitophagy-related proteins, such as PINK1, parkin, NIX (DCT-1 in C. elegans) and BNIP3 (refs. 66,203). Supplementation with NAD+ precursors, such as nicotinamide (NAM), nicotinamide riboside (NR) or nicotinamide mononucleotide (NMN), can increase lifespan and/or improve healthspan in worms, flies and mice111,204,205,206. NAD+ augmentation also prevents memory loss in both amyloid β and tau C. elegans and mouse models of AD, in a mitophagy-dependent manner (requiring pink-1, pdr-1 or dct-1)138. Over seven human clinical trials have shown the safety and bioavailability of NR (1–2 g per day for up to 3 months); there are more than 30 ongoing clinical trials on the use of NR to treat premature aging and other age-related diseases (see a review in ref. 185). Another clinically promising mitophagy inducer is urolithin A, a metabolite of ellagitannins from the gut microflora. Urolithin A extends healthspan and lifespan in C. elegans, with lifespan extension depending on genes involved in autophagy (that is, bec-1, sqst-1 and vps-34) and mitophagy (pink-1, dct-1 and the nonspecific skn-1)207. Intriguingly, urolithin A inhibits memory loss in both amyloid β and tau C. elegans and mouse models of AD in a mitophagy-dependent manner (dependent on pink-1, pdr-1 or dct-1)138. Urolithin A (500 mg and 1,000 mg per day for 4 weeks) was also shown to be safe in a phase 1 clinical trial208. A summary of different lifespan/health-benefit mitophagy inducers can be found in Table 2. Encouraged by the clinical safety of NR and urolithin A, their effects on healthspan and lifespan in older individuals deserve further investigation. Despite recent progress in the identification of novel as well as well-known autophagy-inducing compounds, it is also of great importance to highlight the pleiotropic effects of these pharmacological interventions and to completely understand the full complement of targets with which they interact to use them safely for therapeutic intervention.
While experimental/empirical evidence indicates that autophagy is defective in older individuals, it is conceivable that exposing individuals to autophagy inducers, dietary restriction and exercise late in life could boost autophagy and result in benefits to tissue function209,210 (Fig. 4a). On the basis of preclinical data, it is presumed that autophagy stimulation (ideally to increase autophagy to the levels observed early in adulthood) may be sufficient to provide benefits (Fig. 4b).
Conclusions and future perspectives
Mounting evidence from studies using laboratory animals, human tissues and related clinical trials supports the concept that (1) there is an age-dependent decline in autophagy, (2) autophagy is a crucial determinant of cellular health and organismal longevity and (3) impairment or imbalance in autophagy promotes pathological aging and disease. Given the broad spectrum of unique properties associated with autophagy, we propose that ‘compromised autophagy’ is a central feature of normal aging. Although the relationship between autophagy and aging is often described as ‘decreased autophagy is detrimental’ and ‘increased autophagy is beneficial’, this may be too simplistic a picture. Instead, long-term health benefits will likely arise from achieving the right balance of autophagy, which itself will depend on tissue and organismal age. For example, in C. elegans, impairing autophagy early in life has a negative effect on longevity, whereas knockdown of a specific subset of autophagy genes in adulthood may have beneficial effects on lifespan59. Similarly, increased autophagy through a hypermorphic allele of atg-5 has differential effects on polyglutamine aggregation in the muscles and neurons of C. elegans211. In flies, a mild increase in autophagy extends lifespan, whereas strongly increasing autophagy shortens lifespan212. It should also be noted that autophagy induction may also result in unwanted effects, such as multiple senescence pathologies59 and resistance to cancer therapy (reviewed in ref. 213). Collectively, these observations suggest that the level of and balance among the different forms of autophagy in each tissue are highly specified for each stage of life, and an understanding of this will be crucial for healthy aging. Thus, while different types of autophagy may influence aging to different extents, a central goal for promoting health will be to find approaches that can fine-tune autophagy to the right levels, at the right time and in the right tissues, to enhance health (Fig. 4). To achieve this, it will be critical to develop novel interventions that allow for controlled delivery of autophagy modulators into specific tissues or cell types at precise stages of life. Such therapeutic strategies could then be administered chronically, acutely or in a pulsed fashion as and when required. Additionally, it may be necessary to specifically induce either general or selective autophagy to provide overall long-term health benefits66. For example, premature aging diseases such as ataxia telangiectasia, xeroderma pigmentosum group A and Cockayne syndrome exhibit increased general autophagy but impaired mitophagy; therefore, specifically stimulating mitophagy, rather than general autophagy, would be the most efficient way to counteract pathological features while avoiding detrimental side effects66.
There are many outstanding questions related to autophagy in aging that need to be addressed. For example, what are the intricate mechanisms that orchestrate distinct autophagic pathways? How is autophagy spatially and temporally regulated, and how does disruption of this regulation suppress or promote disease? Are some aspects of autophagy more important in an age- and/or tissue-dependent manner? What are the determining factors that dictate the route of degradation, via the UPS or autophagy? How are clearance mechanisms balanced with synthesis and folding through the proteostasis network? What are the thresholds of life-beneficial and life-detrimental autophagy? In line with the traditional Chinese yin–yang philosophy, autophagy must be balanced, as diminished autophagy results in the accumulation of toxic subcellular components while excessive autophagy can lead to organ atrophy and other detrimental effects14,37,59,69,212. Furthermore, compensatory responses between proteolytic systems (for example, between autophagy and the UPS214) have a critical role in determining the onset and rate of age-related tissue deterioration and should be considered in future experimental designs and data interpretation. Finally, are there any conditions or diseases where we should be cautious about inducing autophagy, in that protection against one form of pathology may increase the risk for another? For example, pancreatic cancer cells may hijack autophagy processes to obtain nutrients for growth; hence, in this condition, autophagy inhibition in combination with cancer chemotherapies may inhibit pancreatic cancer growth215,216. Addressing these questions will facilitate understanding of the aging process and, more importantly, enable identification of novel targets that may be manipulated for therapeutic intervention in age-associated diseases.
References
Leidal, A. M., Levine, B. & Debnath, J. Autophagy and the cell biology of age-related disease. Nat. Cell Biol. 20, 1338–1348 (2018).
Partridge, L., Deelen, J. & Slagboom, P. E. Facing up to the global challenges of ageing. Nature 561, 45–56 (2018).
Fang, E. F. et al. A research agenda for ageing in China in the 21st century (2nd edition): focusing on basic and translational research, long-term care, policy and social networks. Ageing Res. Rev. 64, 101174 (2020).
Dikic, I. & Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018).
Ashford, T. P. & Porter, K. R. Cytoplasmic components in hepatic cell lysosomes. J. Cell Biol. 12, 198–202 (1962).
Deter, R. L. & De Duve, C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J. Cell Biol. 33, 437–449 (1967).
Klionsky, D. J. Autophagy revisited: a conversation with Christian de Duve. Autophagy 4, 740–743 (2008).
Tsukada, M. & Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174 (1993).
Harding, T. M., Morano, K. A., Scott, S. V. & Klionsky, D. J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131, 591–602 (1995).
Thumm, M. et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349, 275–280 (1994).
Schlumpberger, M. et al. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J. Bacteriol. 179, 1068–1076 (1997).
Straub, M., Bredschneider, M. & Thumm, M. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J. Bacteriol. 179, 3875–3883 (1997).
Kaushik, S. & Cuervo, A. M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 (2018).
Hansen, M., Rubinsztein, D. C. & Walker, D. W. Autophagy as a promoter of longevity: insights from model organisms. Nat. Rev. Mol. Cell Biol. 19, 579–593 (2018).
Bjorkoy, G. et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 (2005).
Pankiv, S. et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007).
Kirkin, V. History of the selective autophagy research: how did it begin and where does it stand today? J. Mol. Biol. 432, 3–27 (2020).
Gatica, D., Lahiri, V. & Klionsky, D. J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20, 233–242 (2018).
Menzies, F. M., Fleming, A. & Rubinsztein, D. C. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 16, 345–357 (2015).
Kim, S. et al. Fisetin stimulates autophagic degradation of phosphorylated tau via the activation of TFEB and Nrf2 transcription factors. Sci. Rep. 6, 24933 (2016).
Medina, D. L. et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17, 288–299 (2015).
Settembre, C. et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 (2012).
El-Houjeiri, L. et al. The transcription factors TFEB and TFE3 link the FLCN–AMPK signaling axis to innate immune response and pathogen resistance. Cell Rep. 26, 3613–3628 (2019).
Cuervo, A. M. & Dice, J. F. How do intracellular proteolytic systems change with age? Front. Biosci. 3, D25–D43 (1998).
Sun, Y. et al. Lysosome activity is modulated by multiple longevity pathways and is important for lifespan extension in C. elegans. eLife https://doi.org/10.7554/eLife.55745 (2020).
Sarkis, G. J., Ashcom, J. D., Hawdon, J. M. & Jacobson, L. A. Decline in protease activities with age in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 45, 191–201 (1988).
Hughes, A. L. & Gottschling, D. E. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 492, 261–265 (2012).
Chang, J. T., Kumsta, C., Hellman, A. B., Adams, L. M. & Hansen, M. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. eLife https://doi.org/10.7554/eLife.18459 (2017).
Wilhelm, T. et al. Neuronal inhibition of the autophagy nucleation complex extends life span in post-reproductive C. elegans. Genes Dev. 31, 1561–1572 (2017).
Simonsen, A. et al. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4, 176–184 (2008).
Kaushik, S. et al. Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Rep. 13, 258–265 (2012).
Ott, C., Konig, J., Hohn, A., Jung, T. & Grune, T. Macroautophagy is impaired in old murine brain tissue as well as in senescent human fibroblasts. Redox Biol. 10, 266–273 (2016).
Triplett, J. C. et al. Age-related changes in the proteostasis network in the brain of the naked mole-rat: implications promoting healthy longevity. Biochim. Biophys. Acta 1852, 2213–2224 (2015).
Yu, Y. et al. The alteration of autophagy and apoptosis in the hippocampus of rats with natural aging-dependent cognitive deficits. Behav. Brain Res. 334, 155–162 (2017).
Lipinski, M. M. et al. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc. Natl Acad. Sci. USA 107, 14164–14169 (2010).
Cheon, S. Y., Kim, H., Rubinsztein, D. C. & Lee, J. E. Autophagy, cellular aging and age-related human diseases. Exp. Neurobiol. 28, 643–657 (2019).
Lou, G. et al. Mitophagy and neuroprotection. Trends Mol. Med. https://doi.org/10.1016/j.molmed.2019.07.002 (2019).
Matecic, M. et al. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet. 6, e1000921 (2010).
Toth, M. L. et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 4, 330–338 (2008).
Kuma, A., Komatsu, M. & Mizushima, N. Autophagy-monitoring and autophagy-deficient mice. Autophagy 13, 1619–1628 (2017).
Lin, X. X. et al. DAF-16/FOXO and HLH-30/TFEB function as combinatorial transcription factors to promote stress resistance and longevity. Nat. Commun. 9, 4400 (2018).
Melendez, A. et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391 (2003).
Lapierre, L. R. et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun. 4, 2267 (2013).
Pyo, J. O. et al. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun. 4, 2300 (2013).
Ulgherait, M., Rana, A., Rera, M., Graniel, J. & Walker, D. W. AMPK modulates tissue and organismal aging in a non-cell-autonomous manner. Cell Rep. 8, 1767–1780 (2014).
Fernandez, A. F. et al. Disruption of the beclin 1–BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558, 136–140 (2018).
Silvestrini, M. J. et al. Nuclear export inhibition enhances HLH-30/TFEB activity, autophagy, and lifespan. Cell Rep. 23, 1915–1921 (2018).
Harrison, D. E. et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395 (2009).
Pareja-Cajiao, M. et al. Age-related impairment of autophagy in cervical motor neurons. Exp. Gerontol. 144, 111193 (2021).
Nettesheim, A. et al. Autophagy in the aging and experimental ocular hypertensive mouse model. Invest. Ophthalmol. Vis. Sci. 61, 31 (2020).
Fernando, R. et al. Age-related maintenance of the autophagy–lysosomal system is dependent on skeletal muscle type. Oxid. Med. Cell. Longev. 2020, 4908162 (2020).
Liang, W. et al. Aging is associated with a decline in Atg9b-mediated autophagosome formation and appearance of enlarged mitochondria in the heart. Aging Cell 19, e13187 (2020).
Kamihara, T. & Murohara, T. Bioinformatics analysis of autophagy–lysosomal degradation in cardiac aging. Geriatr. Gerontol. Int. 21, 108–115 (2021).
Fang, Y. et al. Autophagy–Sirt3 axis decelerates hematopoietic aging. Aging Cell 19, e13232 (2020).
Alsaleh, G. et al. Autophagy in T cells from aged donors is maintained by spermidine and correlates with function and vaccine responses. eLife https://doi.org/10.7554/eLife.57950 (2020).
De Risi, M. et al. Mechanisms by which autophagy regulates memory capacity in ageing. Aging Cell 19, e13189 (2020).
Yamamuro, T. et al. Age-dependent loss of adipose Rubicon promotes metabolic disorders via excess autophagy. Nat. Commun. 11, 4150 (2020).
Zhou, B. et al. Mitochondrial permeability uncouples elevated autophagy and lifespan extension. Cell 177, 299–314 (2019).
Ezcurra, M. et al. C. elegans eats its own intestine to make yolk leading to multiple senescent pathologies. Curr. Biol. 28, 2544–2556 (2018).
Lapierre, L. R. et al. Autophagy genes are required for normal lipid levels in C. elegans. Autophagy 9, 278–286 (2013).
Hansen, M. et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 4, e24 (2008).
Labbadia, J. & Morimoto, R. I. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 (2015).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Ravikumar, B., Duden, R. & Rubinsztein, D. C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 11, 1107–1117 (2002).
Ravikumar, B. et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36, 585–595 (2004).
Fang, E. F. et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD+/SIRT1 reduction. Cell 157, 882–896 (2014).
Boland, B. et al. Promoting the clearance of neurotoxic proteins in neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 17, 660–688 (2018).
Fang, E. F. et al. Nuclear DNA damage signalling to mitochondria in ageing. Nat. Rev. Mol. Cell Biol. 17, 308–321 (2016).
Rubinsztein, D. C., Marino, G. & Kroemer, G. Autophagy and aging. Cell 146, 682–695 (2011).
Kumsta, C., Chang, J. T., Schmalz, J. & Hansen, M. Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat. Commun. 8, 14337 (2017).
Juhasz, G., Erdi, B., Sass, M. & Neufeld, T. P. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 21, 3061–3066 (2007).
Hara, T. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006).
Komatsu, M. et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 (2006).
Friedman, L. G. et al. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of α-synuclein and LRRK2 in the brain. J. Neurosci. 32, 7585–7593 (2012).
Schneider, J. L. et al. Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell 14, 249–264 (2015).
Kumsta, C. et al. The autophagy receptor p62/SQST-1 promotes proteostasis and longevity in C. elegans by inducing autophagy. Nat. Commun. 10, 5648 (2019).
Aparicio, R., Rana, A. & Walker, D. W. Upregulation of the autophagy adaptor p62/SQSTM1 prolongs health and lifespan in middle-aged Drosophila. Cell Rep. 28, 1029–1040 (2019).
Demontis, F. & Perrimon, N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813–825 (2010).
Nezis, I. P. et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 180, 1065–1071 (2008).
Lopez, A. et al. A152T tau allele causes neurodegeneration that can be ameliorated in a zebrafish model by autophagy induction. Brain 140, 1128–1146 (2017).
Rocchi, A. et al. A Becn1 mutation mediates hyperactive autophagic sequestration of amyloid oligomers and improved cognition in Alzheimer’s disease. PLoS Genet. 13, e1006962 (2017).
Fu, H. et al. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nat. Neurosci. 22, 47–56 (2019).
Schinaman, J. M., Rana, A., Ja, W. W., Clark, R. I. & Walker, D. W. Rapamycin modulates tissue aging and lifespan independently of the gut microbiota in Drosophila. Sci. Rep. 9, 7824 (2019).
Berger, Z. et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 15, 433–442 (2006).
Audesse, A. J. et al. FOXO3 directly regulates an autophagy network to functionally regulate proteostasis in adult neural stem cells. PLoS Genet. 15, e1008097 (2019).
Leeman, D. S. et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science 359, 1277–1283 (2018).
Garcia-Prat, L. et al. Autophagy maintains stemness by preventing senescence. Nature 529, 37–42 (2016).
Kuma, A. et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004).
Yang, Z., Huang, J., Geng, J., Nair, U. & Klionsky, D. J. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell 17, 5094–5104 (2006).
Suzuki, S. W., Onodera, J. & Ohsumi, Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS ONE 6, e17412 (2011).
Ha, J., Guan, K. L. & Kim, J. AMPK and autophagy in glucose/glycogen metabolism. Mol. Aspects Med. 46, 46–62 (2015).
Adeva-Andany, M. M., Gonzalez-Lucan, M., Donapetry-Garcia, C., Fernandez-Fernandez, C. & Ameneiros-Rodriguez, E. Glycogen metabolism in humans. BBA Clin. 5, 85–100 (2016).
Yao, W. et al. Atg11 is required for initiation of glucose starvation-induced autophagy. Autophagy https://doi.org/10.1080/15548627.2020.1719724 (2020).
Jiang, S., Wells, C. D. & Roach, P. J. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem. Biophys. Res. Commun. 413, 420–425 (2011).
Weber, C. A. et al. β-Oxidation and autophagy are critical energy providers during acute glucose depletion in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 117, 12239–12248 (2020).
Kim, K. A. et al. High glucose condition induces autophagy in endothelial progenitor cells contributing to angiogenic impairment. Biol. Pharm. Bull. 37, 1248–1252 (2014).
Kishmani, P. S. et al. Pompe disease diagnosis and management guideline. Genet. Med. 8, 267–288 (2006).
Kishnani, P. S. et al. Recombinant human acid α-glucosidase: major clinical benefits in infantile-onset Pompe disease. Neurology 68, 99–109 (2007).
Duran, J., Gruart, A., Garcia-Rocha, M., Delgado-Garcia, J. M. & Guinovart, J. J. Glycogen accumulation underlies neurodegeneration and autophagy impairment in Lafora disease. Hum. Mol. Genet. 23, 3147–3156 (2014).
Singh, R. et al. Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009).
Ward, C. et al. Autophagy, lipophagy and lysosomal lipid storage disorders. Biochim. Biophys. Acta 1861, 269–284 (2016).
Kaushik, S. & Cuervo, A. M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 17, 759–770 (2015).
Goeritzer, M. et al. Active autophagy but not lipophagy in macrophages with defective lipolysis. Biochim. Biophys. Acta 1851, 1304–1316 (2015).
Kiffin, R. et al. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J. Cell Sci. 120, 782–791 (2007).
Palikaras, K. et al. Ectopic fat deposition contributes to age-associated pathology in Caenorhabditis elegans. J. Lipid Res. 58, 72–80 (2017).
Stranks, A. J. et al. Autophagy controls acquisition of aging features in macrophages. J. Innate Immun. 7, 375–391 (2015).
Lapierre, L. R., Melendez, A. & Hansen, M. Autophagy links lipid metabolism to longevity in C. elegans. Autophagy 8, 144–146 (2012).
Folick, A. et al. Lysosomal signaling molecules regulate longevity in Caenorhabditis elegans. Science 347, 83–86 (2015).
Lapierre, L. R., Gelino, S., Melendez, A. & Hansen, M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 21, 1507–1514 (2011).
Zhang, T. et al. SIRT3 promotes lipophagy and chaperon-mediated autophagy to protect hepatocytes against lipotoxicity. Cell Death Differ. 27, 329–344 (2020).
Fang, E. F. et al. NAD+ augmentation restores mitophagy and limits accelerated aging in Werner syndrome. Nat. Commun. 10, 5284 (2019).
Baur, J. A. et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444, 337–342 (2006).
Ding, W. X., Li, M. & Yin, X. M. Selective taste of ethanol-induced autophagy for mitochondria and lipid droplets. Autophagy 7, 248–249 (2011).
Kounakis, K., Chaniotakis, M., Markaki, M. & Tavernarakis, N. Emerging roles of lipophagy in health and disease. Front. Cell Dev. Biol. 7, 185 (2019).
Chao, X. et al. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology 155, 865–879 (2018).
Hernandez-Gea, V. et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology 142, 938–946 (2012).
Riffelmacher, T. et al. Autophagy-dependent generation of free fatty acids is critical for normal neutrophil differentiation. Immunity 47, 466–480 (2017).
Kawane, K., Motani, K. & Nagata, S. DNA degradation and its defects. Cold Spring Harb. Perspect. Biol. https://doi.org/10.1101/cshperspect.a016394 (2014).
Houseley, J. & Tollervey, D. The many pathways of RNA degradation. Cell 136, 763–776 (2009).
Buchan, J. R., Kolaitis, R. M., Taylor, J. P. & Parker, R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474 (2013).
Guo, H. et al. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat. Commun. 5, 5276 (2014).
Fujiwara, Y. et al. Direct uptake and degradation of DNA by lysosomes. Autophagy 9, 1167–1171 (2013).
Fujiwara, Y. et al. Discovery of a novel type of autophagy targeting RNA. Autophagy 9, 403–409 (2013).
Aizawa, S. et al. Lysosomal membrane protein SIDT2 mediates the direct uptake of DNA by lysosomes. Autophagy 13, 218–222 (2017).
Aizawa, S. et al. Lysosomal putative RNA transporter SIDT2 mediates direct uptake of RNA by lysosomes. Autophagy 12, 565–578 (2016).
Sliter, D. A. et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262 (2018).
West, A. P. et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015).
Dan, X. et al. DNA damage invokes mitophagy through a pathway involving Spata18. Nucleic Acids Res. 48, 6611–6623 (2020).
Hopfner, K. P. & Hornung, V. Molecular mechanisms and cellular functions of cGAS–STING signalling. Nat. Rev. Mol. Cell Biol. 21, 501–521 (2020).
Johansen, T. & Lamark, T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 432, 80–103 (2020).
Pickles, S., Vigie, P. & Youle, R. J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185 (2018).
Le Guerroue, F. et al. Autophagosomal content profiling reveals an LC3C-dependent piecemeal mitophagy pathway. Mol. Cell 68, 786–796 (2017).
McLelland, G. L., Lee, S. A., McBride, H. M. & Fon, E. A. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J. Cell Biol. 214, 275–291 (2016).
Melentijevic, I. et al. C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature 542, 367–371 (2017).
Nicolas-Avila, J. A. et al. A network of macrophages supports mitochondrial homeostasis in the heart. Cell 183, 94–109 (2020).
Cornelissen, T. et al. Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. eLife https://doi.org/10.7554/eLife.35878 (2018).
Sun, N. et al. Measuring in vivo mitophagy. Mol. Cell 60, 685–696 (2015).
Fang, E. F. et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 22, 401–412 (2019).
Pickrell, A. M. & Youle, R. J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 85, 257–273 (2015).
Palikaras, K., Lionaki, E. & Tavernarakis, N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525–528 (2015).
McWilliams, T. G. et al. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 27, 439–449 (2018).
Fang, E. F. et al. Tomatidine enhances lifespan and healthspan in C. elegans through mitophagy induction via the SKN-1/Nrf2 pathway. Sci. Rep. 7, 46208 (2017).
Du, F. et al. PINK1 signalling rescues amyloid pathology and mitochondrial dysfunction in Alzheimer’s disease. Brain 140, 3233–3251 (2017).
Fang, E. F. et al. NAD+ replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 24, 566–581 (2016).
Mochida, K. et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522, 359–362 (2015).
Hubner, C. A. & Dikic, I. ER-phagy and human diseases. Cell Death Differ. 27, 833–842 (2020).
Park, Y. E. et al. Autophagic degradation of nuclear components in mammalian cells. Autophagy 5, 795–804 (2009).
Dou, Z. et al. Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 (2015).
Papadopoulos, C., Kravic, B. & Meyer, H. Repair or lysophagy: dealing with damaged lysosomes. J. Mol. Biol. 432, 231–239 (2020).
Li, Y. et al. The lysosomal membrane protein SCAV-3 maintains lysosome integrity and adult longevity. J. Cell Biol. 215, 167–185 (2016).
Vesosky, B. & Turner, J. The influence of age on immunity to infection with Mycobacterium tuberculosis. Immunol. Rev. 205, 229–243 (2005).
Papadopoulos, C. & Meyer, H. Detection and clearance of damaged lysosomes by the endo-lysosomal damage response and lysophagy. Curr. Biol. 27, R1330–R1341 (2017).
Gomez-Sintes, R., Ledesma, M. D. & Boya, P. Lysosomal cell death mechanisms in aging. Ageing Res. Rev. 32, 150–168 (2016).
Reggio, A., Buonomo, V. & Grumati, P. Eating the unknown: xenophagy and ER-phagy are cytoprotective defenses against pathogens. Exp. Cell. Res. 396, 112276 (2020).
Levine, B. Eating oneself and uninvited guests: autophagy-related pathways in cell defense. Cell 120, 159–162 (2005).
Rikihisa, Y. Glycogen autophagosomes in polymorphonuclear leukocytes induced by rickettsiae. Anat. Rec. 208, 319–327 (1984).
Rich, K. A., Burkett, C. & Webster, P. Cytoplasmic bacteria can be targets for autophagy. Cell. Microbiol. 5, 455–468 (2003).
Nakagawa, I. et al. Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040 (2004).
Gutierrez, M. G. et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004).
Kimmey, J. M. & Stallings, C. L. Bacterial pathogens versus autophagy: implications for therapeutic interventions. Trends Mol. Med. 22, 1060–1076 (2016).
Upadhyay, S., Mittal, E. & Philips, J. A. Tuberculosis and the art of macrophage manipulation. Pathog. Dis. https://doi.org/10.1093/femspd/fty037 (2018).
Watson, R. O. et al. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe 17, 811–819 (2015).
Franco, L. H. et al. The ubiquitin ligase Smurf1 functions in selective autophagy of Mycobacterium tuberculosis and anti-tuberculous host defense. Cell Host Microbe 21, 59–72 (2017).
Bah, A. & Vergne, I. Macrophage autophagy and bacterial infections. Front. Immunol. 8, 1483 (2017).
Jayaswal, S. et al. Identification of host-dependent survival factors for intracellular Mycobacterium tuberculosis through an siRNA screen. PLoS Pathog. 6, e1000839 (2010).
Kim, J. J. et al. Host cell autophagy activated by antibiotics is required for their effective antimycobacterial drug action. Cell Host Microbe 11, 457–468 (2012).
Watson, R. O., Manzanillo, P. S. & Cox, J. S. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 150, 803–815 (2012).
Wang, J. et al. MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog. 9, e1003697 (2013).
Liang, X. H. et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 72, 8586–8596 (1998).
Orvedahl, A. et al. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7, 115–127 (2010).
Orvedahl, A. et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1, 23–35 (2007).
Mijaljica, D. & Klionsky, D. J. Autophagy/virophagy: a “disposal strategy” to combat COVID-19. Autophagy https://doi.org/10.1080/15548627.2020.1782022 (2020).
Shojaei, S., Suresh, M., Klionsky, D. J., Labouta, H. I. & Ghavami, S. Autophagy and SARS-CoV-2 infection: a possible smart targeting of the autophagy pathway. Virulence 11, 805–810 (2020).
Carmona-Gutierrez, D. et al. Digesting the crisis: autophagy and coronaviruses. Microb. Cell 7, 119–128 (2020).
Choi, J. et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 40, 924–935 (2014).
Ghartey-Kwansah, G. et al. Autophagy in the control and pathogenesis of parasitic infections. Cell Biosci. 10, 101 (2020).
O’Driscoll, M. et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature https://doi.org/10.1038/s41586-020-2918-0 (2020).
Gelino, S. et al. Intestinal autophagy improves healthspan and longevity in C. elegans during dietary restriction. PLoS Genet. 12, e1006135 (2016).
Minnerly, J., Zhang, J., Parker, T., Kaul, T. & Jia, K. The cell non-autonomous function of ATG-18 is essential for neuroendocrine regulation of Caenorhabditis elegans lifespan. PLoS Genet. 13, e1006764 (2017).
Bai, H., Kang, P., Hernandez, A. M. & Tatar, M. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet. 9, e1003941 (2013).
Carnio, S. et al. Autophagy impairment in muscle induces neuromuscular junction degeneration and precocious aging. Cell Rep. 8, 1509–1521 (2014).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013).
Dong, S. et al. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature 591, 117–123 (2021).
Bourdenx, M. et al. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell 184, 2696–2714 (2021).
Lautrup, S., Sinclair, D. A., Mattson, M. P. & Fang, E. F. NAD+ in brain aging and neurodegenerative disorders. Cell Metab. 30, 630–655 (2019).
Franceschi, C., Garagnani, P., Parini, P., Giuliani, C. & Santoro, A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14, 576–590 (2018).
Medzhitov, R. Origin and physiological roles of inflammation. Nature 454, 428–435 (2008).
Riffelmacher, T., Richter, F. C. & Simon, A. K. Autophagy dictates metabolism and differentiation of inflammatory immune cells. Autophagy 14, 199–206 (2018).
Swanson, K. V., Deng, M. & Ting, J. P. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489 (2019).
Sun, Q., Fan, J., Billiar, T. R. & Scott, M. J. Inflammasome and autophagy regulation—a two-way street. Mol. Med. 23, 188–195 (2017).
Menzies, F. M. et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron 93, 1015–1034 (2017).
Rubinsztein, D. C., Codogno, P. & Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 11, 709–730 (2012).
Wood, J. G. et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430, 686–689 (2004).
Morselli, E. et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 192, 615–629 (2011).
Pietrocola, F. et al. Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ. 22, 509–516 (2015).
Madeo, F., Eisenberg, T., Pietrocola, F. & Kroemer, G. Spermidine in health and disease. Science https://doi.org/10.1126/science.aan2788 (2018).
Eisenberg, T. et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 22, 1428–1438 (2016).
Zhang, H. et al. Polyamines control eIF5A hypusination, TFEB translation, and autophagy to reverse B cell senescence. Mol. Cell 76, 110–125 (2019).
Eisenberg, T. et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 11, 1305–1314 (2009).
Song, H. L., Demirev, A. V., Kim, N. Y., Kim, D. H. & Yoon, S. Y. Ouabain activates transcription factor EB and exerts neuroprotection in models of Alzheimer’s disease. Mol. Cell. Neurosci. 95, 13–24 (2019).
Huang, R. et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 57, 456–466 (2015).
Lee, I. H. et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl Acad. Sci. USA 105, 3374–3379 (2008).
Fang, E. F. Mitophagy and NAD+ inhibit Alzheimer disease. Autophagy 15, 1112–1114 (2019).
Mouchiroud, L. et al. The NAD+/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154, 430–441 (2013).
Zhang, H. et al. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443 (2016).
Mitchell, S. J. et al. Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Metab. 27, 667–676 (2018).
Ryu, D. et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat. Med. 22, 879–888 (2016).
Andreux, P. A. et al. The mitophagy activator urolithin A is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Nat. Metab. 1, 593–603 (2019).
Escobar, K. A., Cole, N. H., Mermier, C. M. & VanDusseldorp, T. A. Autophagy and aging: maintaining the proteome through exercise and caloric restriction. Aging Cell 18, e12876 (2019).
de Cabo, R. & Mattson, M. P. Effects of intermittent fasting on health, aging, and disease. N. Engl. J. Med. 381, 2541–2551 (2019).
Alexander-Floyd, J. et al. Unexpected cell type-dependent effects of autophagy on polyglutamine aggregation revealed by natural genetic variation in C. elegans. BMC Biol. 18, 18 (2020).
Bjedov, I. et al. Fine-tuning autophagy maximises lifespan and is associated with changes in mitochondrial gene expression in Drosophila. PLoS Genet. 16, e1009083 (2020).
Mulcahy Levy, J. M. & Thorburn, A. Autophagy in cancer: moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ. 27, 843–857 (2020).
Park, C. & Cuervo, A. M. Selective autophagy: talking with the UPS. Cell Biochem. Biophys. 67, 3–13 (2013).
Yang, S. et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25, 717–729 (2011).
Piffoux, M., Eriau, E. & Cassier, P. A. Autophagy as a therapeutic target in pancreatic cancer. Br. J. Cancer 124, 333–344 (2021).
Matsuura, A., Tsukada, M., Wada, Y. & Ohsumi, Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192, 245–250 (1997).
Alvers, A. L. et al. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy 5, 847–849 (2009).
Rana, A. et al. Promoting Drp1-mediated mitochondrial fission in midlife prolongs healthy lifespan of Drosophila melanogaster. Nat. Commun. 8, 448 (2017).
Velikkakath, A. K., Nishimura, T., Oita, E., Ishihara, N. & Mizushima, N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell 23, 896–909 (2012).
Xu, P. et al. Atg2, Atg9 and Atg18 in mitochondrial integrity, cardiac function and healthspan in Drosophila. J. Mol. Cell. Cardiol. 127, 116–124 (2019).
Maruyama, T. & Noda, N. N. Autophagy-regulating protease Atg4: structure, function, regulation and inhibition. J. Antibiot. https://doi.org/10.1038/ja.2017.104 (2017).
Yang, J. et al. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age 35, 11–22 (2013).
Reggiori, F. & Klionsky, D. J. Autophagosomes: biogenesis from scratch? Curr. Opin. Cell Biol. 17, 415–422 (2005).
Ruckenstuhl, C. et al. Lifespan extension by methionine restriction requires autophagy-dependent vacuolar acidification. PLoS Genet. 10, e1004347 (2014).
Bjedov, I. et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11, 35–46 (2010).
Jia, K. & Levine, B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy 3, 597–599 (2007).
Hars, E. S. et al. Autophagy regulates ageing in C. elegans. Autophagy 3, 93–95 (2007).
He, C. & Klionsky, D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 (2009).
Walczak, M. & Martens, S. Dissecting the role of the Atg12–Atg5–Atg16 complex during autophagosome formation. Autophagy 9, 424–425 (2013).
Teter, S. A. et al. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 276, 2083–2087 (2001).
Tang, F. et al. A life-span extending form of autophagy employs the vacuole–vacuole fusion machinery. Autophagy 4, 874–886 (2008).
Rieter, E. et al. Atg18 function in autophagy is regulated by specific sites within its β-propeller. J. Cell Sci. 126, 593–604 (2013).
McQuary, P. R. et al. C. elegans S6K mutants require a creatine-kinase-like effector for lifespan extension. Cell Rep. 14, 2059–2067 (2016).
Narendra, D., Tanaka, A., Suen, D. F. & Youle, R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008).
Rana, A., Rera, M. & Walker, D. W. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proc. Natl Acad. Sci. USA 110, 8638–8643 (2013).
Zhuang, N., Li, L., Chen, S. & Wang, T. PINK1-dependent phosphorylation of PINK1 and parkin is essential for mitochondrial quality control. Cell Death Dis. 7, e2501 (2016).
Schiavi, A. et al. Iron-starvation-induced mitophagy mediates lifespan extension upon mitochondrial stress in C. elegans. Curr. Biol. 25, 1810–1822 (2015).
Johansen, T. & Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 (2011).
Parekh, V. V. et al. Autophagy-related protein Vps34 controls the homeostasis and function of antigen cross-presenting CD8α+ dendritic cells. Proc. Natl Acad. Sci. USA 114, E6371–E6380 (2017).
Cabreiro, F. et al. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153, 228–239 (2013).
Partridge, L., Fuentealba, M. & Kennedy, B. K. The quest to slow ageing through drug discovery. Nat. Rev. Drug Discov. 19, 513–532 (2020).
Lu, Y. X. et al. A TORC1-histone axis regulates chromatin organisation and non-canonical induction of autophagy to ameliorate ageing. eLife 10, e62233 (2021).
Fivenson, E. M. et al. Mitophagy in neurodegeneration and aging. Neurochem. Int. 109, 202–209 (2017).
Honda, Y., Tanaka, M. & Honda, S. Trehalose extends longevity in the nematode Caenorhabditis elegans. Aging Cell 9, 558–569 (2010).
Shi, D. et al. The precursor of PI(3,4,5)P3 alleviates aging by activating daf-18(Pten) and independent of daf-16. Nat. Commun. 11, 4496 (2020).
Acknowledgements
E.F.F. was supported by HELSE SØR-ØST (2017056, 2020001 and 2021021), the Research Council of Norway (262175 and 277813), the National Natural Science Foundation of China (81971327), Akershus University Hospital (269901 and 261973), the Civitan Norges Forskningsfond for Alzheimers sykdom (for a three-year PhD fellowship, 281931), the Czech Republic–Norway KAPPA programme (with M. Vyhnálek, TO01000215) and the Rosa sløyfe/Norwegian Cancer Society & Norwegian Breast Cancer Society (207819). J.L. is supported by a BBSRC David Phillips Fellowship (BB/P005535/1), a BBSRC Responsive Mode Grant (BB/T013273/1), an AMS Springboard Award (SBF004\1051) and a Wellcome Trust ISSF award (ISSF3/H17RCO/NG18). A.K.S. is supported by Wellcome Trust Investigator Award 103830/Z/14/Z. A.S. is supported by the Norwegian Cancer Society (171318) and the Research Council of Norway (221831) and its Centres of Excellence funding scheme (262652), as well as by HELSE SØR-ØST (2020032). T.J. is supported by the Norwegian Cancer Society (190214) and the Research Council of Norway (249884). N.T. was supported by grants from the European Research Council (ERC GA695190, MANNA; ERC-GA737599, NeuronAgeScreen). M.H. is supported by the National Institutes of Health (AG038664 and GM117466) as well as by the American Federation for Aging Research and the Larry L. Hillblom Foundation. R.M. is supported by the National Institutes of Health (AG057296 and AG054407), the Tau Consortium and the Daniel F. and Ada L. Rice Foundation. D.C.R. is supported by the UK Dementia Research Institute (funded by the MRC), Alzheimer’s Research UK and the Alzheimer’s Society (UKDRI-2002 to D.C.R.), the Tau Consortium, Alzheimer’s Research UK, an anonymous donation to the Cambridge Centre for Parkinson-Plus and the Roger de Spoelberch Prize. I.B. is supported by funding from the European Research Council (ERC PoC 842174) and the Cancer Research UK City of London Centre Award (C7893/A28990). L.P. is supported by a Wellcome Trust Collaborative Award, the Max Planck Society and the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement number 268739. G.K. is supported by the Ligue contre le Cancer (équipe labellisée); Agence National de la Recherche (ANR) – Projets blancs; AMMICa US23/CNRS UMS3655; Association pour la recherche sur le cancer (ARC); Association “Ruban Rose”; Cancéropôle Ile-de-France; Fondation pour la Recherche Médicale (FRM); a donation by Elior; Equipex Onco-Pheno-Screen; European Joint Programme on Rare Diseases (EJPRD); Gustave Roussy Odyssea, the European Union Horizon 2020 Projects Oncobiome and Crimson; Fondation Carrefour; Institut National du Cancer (INCa); Inserm (HTE); Institut Universitaire de France; LabEx Immuno-Oncology (ANR-18-IDEX-0001); the Leducq Foundation; the RHU Torino Lumière; Seerave Foundation; SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); and SIRIC Cancer Research and Personalized Medicine (CARPEM). This study contributes to the IdEx Université de Paris ANR-18-IDEX-0001. All figures were created with BioRender.com.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
E.F.F. has a CRADA arrangement with ChromaDex and is a consultant to Aladdin Healthcare Technologies, the Vancouver Dementia Prevention Centre and Intellectual Labs. A.K.S. is a consultant to Oxford Healthspan. D.C.R. is a consultant for Aladdin Healthcare Technologies, Drishti Discoveries and Nido Biosciences. G.K. is a scientific co-founder of everImmune, Samsara Therapeutics and Therafast Bio. All other authors declare no competing interests.
Additional information
Peer review information Nature Aging thanks Louis Lapierre, Esther Wong and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1.
Rights and permissions
About this article
Cite this article
Aman, Y., Schmauck-Medina, T., Hansen, M. et al. Autophagy in healthy aging and disease. Nat Aging 1, 634–650 (2021). https://doi.org/10.1038/s43587-021-00098-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-021-00098-4
This article is cited by
-
Autophagy dysfunction contributes to NLRP1 inflammasome-linked depressive-like behaviors in mice
Journal of Neuroinflammation (2024)
-
The application of nanoparticles-based ferroptosis, pyroptosis and autophagy in cancer immunotherapy
Journal of Nanobiotechnology (2024)
-
Neuroactive steroid effects on autophagy in a human embryonic kidney 293 (HEK) cell model
Scientific Reports (2024)
-
Reduction of DHHC5-mediated beclin 1 S-palmitoylation underlies autophagy decline in aging
Nature Structural & Molecular Biology (2024)
-
Inhibition of S6K lowers age-related inflammation and increases lifespan through the endolysosomal system
Nature Aging (2024)