Abstract
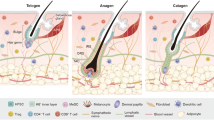
Regressive changes in epithelial stem cells underlie mammalian skin aging, but the driving mechanisms are not well understood. Here, we report that mouse skin hair follicle stem cell (HFSC) aging is initiated by their intrinsic upregulation of miR-31, a microRNA that can be induced by physical injury or genotoxic stress and is also strongly upregulated in aged human skin epithelium. Using transgenic and conditional knockout mouse models plus a lineage-tracing technique, we show that miR-31 acts as a key driver of HFSC aging by directly targeting Clock, a core circadian clock gene whose deregulation activates a MAPK/ERK cascade to induce HFSC depletion via transepidermal elimination. Notably, blocking this pathway by either conditional miR-31 ablation or clinically approved MAPK/ERK inhibitors provides safe and effective protection against skin aging, enlightening a promising therapeutic avenue for treating skin aging and other genotoxic stress-induced skin conditions such as radiodermatitis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
miRNA target predictions were downloaded from the TargetScan website http://www.targetscan.org/mmu_71/. RNA-seq data have been deposited at Bio-Med Big Data Center with accession number OEP000965. A copy was also deposited to NCBI Sequence Read Archive (SRA) as SRP326041. Other related raw data files, the so-called ‘minimum dataset’, are deposited at https://www.biosino.org/node/project/detail/OEP002383 and are publicly available.
References
Goodell, M. A. & Rando, T. A. Stem cells and healthy aging. Science 350, 1199–1204 (2015).
Ermolaeva, M., Neri, F., Ori, A. & Rudolph, K. L. Cellular and epigenetic drivers of stem cell ageing. Nat. Rev. Mol. Cell Biol. 19, 594–610 (2018).
Blanpain, C. & Fuchs, E. Epidermal stem cells of the skin. Annu. Rev. Cell Dev. Biol. 22, 339–373 (2006).
Rittié, L. & Fisher, G. J. Natural and sun-induced aging of human skin. Cold Spring Harb. Perspect. Med. 5, a015370 (2015).
Fuchs, E. Skin stem cells: rising to the surface. J. Cell Biol. 180, 273–284 (2008).
Alonso, L. & Fuchs, E. The hair cycle. J. Cell Sci. 119, 391–393 (2006).
Ito, M. et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11, 1351–1354 (2005).
Ge, Y. et al. Stem cell lineage infidelity drives wound repair and cancer. Cell 169, 636–650 (2017).
Oh, J., Lee, Y. D. & Wagers, A. J. Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat. Med. 20, 870–880 (2014).
Vijg, J. & Suh, Y. Genome instability and aging. Annu. Rev. Physiol. 75, 645–668 (2013).
Garinis, G. A., van der Horst, G. T., Vijg, J. & Hoeijmakers, J. H. DNA damage and ageing: new-age ideas for an age-old problem. Nat. Cell Biol. 10, 1241–1247 (2008).
Bertell, R. X-ray exposure and premature aging. J. Surg. Oncol. 9, 379–391 (1977).
Matsumura, H. et al. Hair follicle aging is driven by transepidermal elimination of stem cells via COL17A1 proteolysis. Science 351, aad4395 (2016).
Yi, R. Concise review: mechanisms of quiescent hair follicle stem cell regulation. Stem Cells 35, 2323–2330 (2017).
Fuchs, E. Skin stem cells in silence, action, and cancer. Stem Cell Rep. 10, 1432–1438 (2018).
Keyes, B. E. & Fuchs, E. Stem cells: aging and transcriptional fingerprints. J. Cell Biol. https://doi.org/10.1083/jcb.201708099 (2018).
Baker, K. W. & Habowsky, J. E. EDTA separation and ATPase Langerhans cell staining in the mouse epidermis. J. Investig. Dermatol. 80, 104–107 (1983).
Singh, M., Alavi, A., Wong, R. & Akita, S. Radiodermatitis: a review of our current understanding. Am. J. Clin. Dermatol. 17, 277–292 (2016).
Xu, Z. et al. Embryonic attenuated Wnt/β-catenin signaling defines niche location and long-term stem cell fate in hair follicle. eLife 4, e10567 (2015).
Panich, U., Sittithumcharee, G., Rathviboon, N. & Jirawatnotai, S. Ultraviolet radiation-induced skin aging: the role of DNA damage and oxidative stress in epidermal stem cell damage mediated skin aging. Stem Cells Int. 2016, 7370642–7370642 (2016).
Zhang, L., Stokes, N., Polak, L. & Fuchs, E. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell 8, 294–308 (2011).
Shi, J. et al. MiR-31 mediates inflammatory signaling to promote re-epithelialization during skin wound healing. J. Invest. Dermatol. 138, 2253–2263 (2018).
Rotty, J. D. & Coulombe, P. A. A wound-induced keratin inhibits Src activity during keratinocyte migration and tissue repair. J. Cell Biol. 197, 381–389 (2012).
Tice, R. R. et al. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ. Mol. Mutagen. 35, 206–221 (2000).
Mah, L. J., El-Osta, A. & Karagiannis, T. C. γH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia 24, 679–686 (2010).
Lee, B. Y. et al. Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging Cell 5, 187–195 (2006).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Friedman, R. C., Farh, K. K.-H., Burge, C. B. & Bartel, D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009).
Lien, W. H. et al. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell 9, 219–232 (2011).
White, A. C. et al. Stem cell quiescence acts as a tumour suppressor in squamous tumours. Nat. Cell Biol. 16, 99–107 (2014).
Sebolt-Leopold, J. S. & Herrera, R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat. Rev. Cancer 4, 937–947 (2004).
Escuin-Ordinas, H., Li, S., Xie, M. W., Sun, L. & Hugo, W. Cutaneous wound healing through paradoxical MAPK activation by BRAF inhibitors. Nat. Commun. https://doi.org/10.1038/ncomms12348 (2016).
Morris, E. J. et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 3, 742–750 (2013).
Tata, S., Weiner, N. & Flynn, G. Relative influence of ethanol and propylene glycol cosolvents on deposition of minoxidil into the skin. J. Pharm. Sci. 83, 1508–1510 (1994).
Janich, P. et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature 480, 209–214 (2011).
Yan, S. et al. NF-κB-induced microRNA-31 promotes epidermal hyperplasia by repressing protein phosphatase 6 in psoriasis. Nat. Commun. 6, 7652 (2015).
Woo, W. M., Atwood, S. X., Zhen, H. H. & Oro, A. E. Rapid genetic analysis of epithelial-mesenchymal signaling during hair regeneration. J. Vis. Exp. https://doi.org/10.3791/4344 (2013).
Zou, Z. et al. A single-cell transcriptomic atlas of human skin aging. Dev. Cell 56, 383–397 (2021).
Deng, M. et al. A role for the mitogen-activated protein kinase kinase kinase 1 in epithelial wound healing. Mol. Biol. Cell 17, 3446–3455 (2006).
Peus, D. et al. UVB activates ERK1/2 and p38 signaling pathways via reactive oxygen species in cultured keratinocytes. J. Invest. Dermatol. 112, 751–756 (1999).
Chernyavsky, A. I., Arredondo, J., Karlsson, E., Wessler, I. & Grando, S. A. The Ras/Raf-1/MEK1/ERK signaling pathway coupled to integrin expression mediates cholinergic regulation of keratinocyte directional migration. J. Biol. Chem. 280, 39220–39228 (2005).
Matsubayashi, Y., Ebisuya, M., Honjoh, S. & Nishida, E. ERK activation propagates in epithelial cell sheets and regulates their migration during wound healing. Curr. Biol. 14, 731–735 (2004).
Lay, K., Kume, T. & Fuchs, E. FOXC1 maintains the hair follicle stem cell niche and governs stem cell quiescence to preserve long-term tissue-regenerating potential. Proc. Natl Acad. Sci. USA 113, E1506–E1515 (2016).
Chen, T. et al. An RNA interference screen uncovers a new molecule in stem cell self-renewal and long-term regeneration. Nature 485, 104–108 (2012).
Hood, S. & Amir, S. The aging clock: circadian rhythms and later life. J. Clin. Invest. 127, 437–446 (2017).
Kondratov, R. V., Kondratova, A. A., Gorbacheva, V. Y., Vykhovanets, O. V. & Antoch, M. P. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 20, 1868–1873 (2006).
Wisor, J. P. et al. Sleep deprivation effects on circadian clock gene expression in the cerebral cortex parallel electroencephalographic differences among mouse strains. J. Neurosci. 28, 7193–7201 (2008).
Möller-Levet, C. S. et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc. Natl Acad. Sci. USA 110, E1132–E1141 (2013).
Tanimura, S. et al. Hair follicle stem cells provide a functional niche for melanocyte stem cells. Cell Stem Cell 8, 177–187 (2011).
Lu, Z. et al. Hair follicle stem cells regulate retinoid metabolism to maintain the self-renewal niche for melanocyte stem cells. eLife https://doi.org/10.7554/eLife.52712 (2020).
Aoki, H., Hara, A., Motohashi, T. & Kunisada, T. Keratinocyte stem cells but not melanocyte stem cells are the primary target for radiation-induced hair graying. J. Invest. Dermatol. 133, 2143–2151 (2013).
Levy, V., Lindon, C., Zheng, Y., Harfe, B. D. & Morgan, B. A. Epidermal stem cells arise from the hair follicle after wounding. FASEB J. 21, 1358–1366 (2007).
Lichti, U., Anders, J. & Yuspa, S. H. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 3, 799–810 (2008).
Beronja, S., Livshits, G., Williams, S. & Fuchs, E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nat. Med. 16, 821–827 (2010).
Campbell, J. J. et al. CCR7 expression and memory T cell diversity in humans. J. Immunol. 166, 877–884 (2001).
Końca, K. et al. A cross-platform public domain PC image-analysis program for the comet assay. Mutation Res. 534, 15–20 (2003).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Varet, H., Brillet-Guéguen, L., Coppée, J. Y. & Dillies, M. A. SARTools: a DESeq2- and EdgeR-based R pipeline for comprehensive differential analysis of RNA-seq data. PLoS ONE 11, e0157022 (2016).
Callahan, B. J., Sankaran, K., Fukuyama, J. A., McMurdie, P. J. & Holmes, S. P. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Res. 5, 1492 (2016).
Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P. & Burge, C. B. Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003).
Keyes, B. E. et al. Impaired epidermal to dendritic T cell signaling slows wound repair in aged skin. Cell 167, 1323–1338 (2016).
Nowak, J. A. & Fuchs, E. Isolation and culture of epithelial stem cells. Methods Mol. Biol. 482, 215–232 (2009).
Ge, Y., Zhang, L., Nikolova, M., Reva, B. & Fuchs, E. Strand-specific in vivo screen of cancer-associated miRNAs unveils a role for miR-21∗ in SCC progression. Nat. Cell Biol. 18, 111–121 (2016).
Elbashir, S. M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001).
Acknowledgements
This study is supported by the National Key R&D Program of China 2017YFA0103500 (to Liang Zhang), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDA160101XX to Liang Zhang), the National Key R&D Program of China 2017YFA0102800 (to Liang Zhang), the National Natural Science Foundation of China (grant nos. 32070730, 31471385 and 81673104 to Liang Zhang), the National Thousand Young Talents Award of China (to Liang Zhang), Shanghai Municipal Science and Technology Major Project (grant no. 2017SHZDZX01 to Liang Zhang), innovative research team of high-level local universities in Shanghai (Liang Zhang), the Clinical Research Plan of SHDC (grant no. SHDC2020CR1019B to Q.L.), Shanghai Municipal Key Clinical Specialty (grant no. shslczdzk00901 to Q.L.), innovative research team of high-level local universities in Shanghai (grant no. SSMU-ZDCX20180700 to Q.L.), the National Natural Science Foundation of China (grant no. 82072191 to C.L.), Shanghai Municipal Key Clinical Specialty (grant no. shslczdzk00901 to C.L.), the China Postdoctoral Science Foundation Grant (grant no. 2018M630449 to C.L.). We thank E. Fuchs (Rockefeller University) for providing K14-rtTA mice, T. Chen (National Institute of Biological Sciences) for providing Sox9-CreER;mTmG mice, J. Han (Xiamen University) for providing human CLOCK cDNA and C. Yang (Changhai Hospital, Second Military Medical University) for critical reading of the manuscript and helpful advice.
Author information
Authors and Affiliations
Contributions
Liang Zhang and Y.Y. designed the experiments and wrote the manuscript. Y.Y. performed most of the experiments. Liang Zhang performed data analysis of RNA-seq and miRNA-seq. X.Z. and F.L assisted with mice works. X.Z., F.L., P.Z, Liping Zhang, Y.P., X.Y., Y.L. and P.H. provided protocols, reagents and scientific advice. C.L. and Q.L. were responsible for the collection and processing of human clinical samples. All authors provided intellectual input and vetted and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
Patents related to this work have been filed to China National Intellectual Property Administration with application nos. 202010074345.7, 202010813757.8, 201911175875.4 and 202010231136.9. The authors declare no other competing interests. Authors on all patent applications are Liang Zhang and Y.Y.
Additional information
Peer review information Nature Aging thanks Yeijing Ge, Rui Yi and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
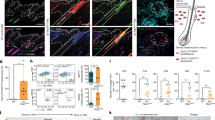
Extended Data Fig. 1 miR-31 upregulation is an early event in both HFSC aging and wound response.
a, Whole-mount IF staining of mouse backskin epithelium isolated by EDTA digestion. VIM: vimentin. Scale bars, 50μm. b, In situ hybridization of young (8 weeks)/old (22 months) backskin sections. Brackets: HF Bulge. Scale bars: 50μm. c, FACS sorting profiles of young (8 weeks)/middle (12 months)/old (22 months) HFSCs (CD34 + CD49fhi, boxed). d, In situ hybridization of young/old backskin sections, samples were collected at 10 days post depilation. Brackets: HF Bulge. Arrow heads: HF matrix. Scale bars: 50μm. e, Oil red staining of same animal Rad/noRad backskin sections at A2. Scale bars: 50 μm. Arrows: SG. f, Left panel: photos of backskin full-thickness wound healing of LIR or Ctrl mice at A1 time. d0-9: days post wounding. Scale bar: 5 mm. Right panel: Quantification of the left healing process. Vertical axis: % of wound healed. P values are for Rad vs noRad, n = 3 biologically independent mice. g, Left panel: white-field images of A1 time backskin exoplant cultures at day 6. Dash lines: borders of migrating keratinocyte sheet (MK sheet). #:originalexoplants. Scale bars: 100μm. Right panel: Quantification of the MK sheet area. Vertical axis: relative % of MK sheet area vs Ctrl-L. n = 6 exoplants separated from 3 mice. h, Left: representative FACS profiles of same animal noRad/Rad skin at A1 time. Box and %: CD49fhiCD34 + HFSCs and its % among CD49f + cells. Right: quantification of relative HFSC % vs noRad. **: p < 0.01, two-tail t-test, n = 4 biologically independent mice. i, Left panel: Skin surface images of same animal noRad/Rad areas at different days (d) after Wax2. Scale bars: 500 μm. Right panel: Quantification of relative hair density in same animal noRad/Rad backskin regions at A2. **: p < 0.01, two-tail t-test, n = 9 samples separated from 3 mice. j, Whole-mount IF staining and HF size quantification of same animal Rad/noRad backskin epithelium at A2. n = 5 HFs. Scale bars: 50 μm. For all relevant figures, data are represented as mean ± SEM. P values were calculated with two tail t-test.
Extended Data Fig. 2 miR-31 up-regulation is an early event in both HFSC aging and wound response.
a, Schematic of the Sox9-CreER;mTmG mouse line and its usage for lineage tracing of HFSC progenies. Tam: Tamoxifen. b, Fluorescent microscope images of Tam-labeled or unlabeled Sox9-CreER;mTmG mice backskin sections at 2 days post labeling, as indicated in the left diagram. Dashed lines outline basement membrane. Scale bars, 50μm. c, GFP fluorescent skin surface images of same animal Rad/noRad skin of Tam-labeled Sox9-CreER;mTmG mice at day 8 post LIR without waxing. The mice were Tam pulse labeled 2 days before LIR. Scale bar, 2 mm. d, Fluorescent microscope images of labeled Sox9-CreER;mTmG mice backskin sections at day 0 (d0), day 1 (d1) or day 2 (d2) post wax in LIR procedure. Scale bars, 50μm. Arrow heads: GFP + HF progenies in IFE. e, Fluorescent microscope images of Sox9-CreER;mTmG mice backskin sections without Tam-labeled at day 28 post wax in LIR procedure. Scale bars, 50μm. f, miRNA qRT-PCR of cultured MKs or backskin epithelium (Epi) 24 hours after 0mJ/40mJ/60mJ UVB pulse. n = 4 biologically independent samples each. g, In situ hybridization images of backskin sections of WT mice at ages indicated. Scale bars: 50μm. Scr/31 indicate scramble/miR-31 in situ probes used. Brackets: HFSC region (bulge). h, IF staining of Wax (W) or Rub(R) backskin sections at different days (d) post treatment. Staining antibodies were used as indicated. Scale bars, 50μm. i, Fluorescent microscope images of labeled Sox9-CreER;mTmG mice backskin sections at day 0 (d0) or day 8 (d8) post Rub, as illustrated in the top diagram. Scale bars, 50μm. Arrow heads: GFP + HF progenies in IFE. j, IF staining images of age 21 female human eyelid skin section (human) and 7 weeks age mouse backskin section (mouse). Scale bars:100μm. For all relevant figures, data are represented as mean ± SEM. P values were calculated with two tail t-test.
Extended Data Fig. 3 miR-31 up-regulation in skin epithelium induces HFSC transepidermal differentiation and aging-like phenotypes.
a, miR-31 qRT-PCR of young/old/WT/DTG backskin epidermis. n = 3 biologically independent mice each. WT/DTG mice were induced by Dox at P48 and samples collected 2 days later. b, Backskin photographs of WT/DTG littermates at age P48 and P62 without Dox induction. Scale bars: 1 cm. c, IF staining images of the above DTG and WT backskin sections. DAPI is in blue. Scale bars: 50 μm. d, IF staining images of backskin sections of P55 WT/DTG littermates (+Dox since P21). Arrow heads: K10 and LOR signals in HF. Scale bars: 50 μm. CD49f is in Red. DAPI is in blue. e, Left diagram: Schematic of full-thickness wound healing assay procedures in Dox induced WT/DTG mice. The mice were Dox induced at P48 and wounded at P50. Middle: Photographs of the wound healing process. Scale bars: 5 mm. Right: Quantification of the wound healing process in E Figure. Vertical axis: % wound healed. n = 6 biologically independent mice. f, Left panel: white-field images of day 4 exoplant cultures of backskin samples from DTG/WT littermates (+Dox since P48). Dashed lines: borders of MK sheet migration. #: original exoplant. Scale bars: 100 μm. Right panel: Quantification of the MK sheet area. n = 6 exoplants separated from 3 mice. g, IF staining images of full-thickness wound d7 backskin sections of P55 WT/DTG littermates (+Dox since P21). Scale bars: 200 μm. Brackets indicates open wound area. For all relevant figures, data are represented as mean ± SEM. P values were calculated with two tail t-test.
Extended Data Fig. 4 miR-31 up-regulation in skin epithelium induces HFSC transepidermal differentiation and aging-like phenotypes.
a, IF staining images of backskin sections of no Wax/post Wax 48 hr WT/DTG littermates. Scale bars: 50μm. b, Backskin surface images of Tam-labeled WT/DTG Sox9-CreER;mTmG littermates after Dox induction and Wax. The mice were waxed at P50 ( + Dox and Tam since P48) and photographed at day 3 post waxing. Scale bars: 2 mm. c, IF staining images of backskin sections of post Wax day0 and day 3 WT/DTG littermates. They were waxed at P50 ( + Dox since P48). Scale bars: 50μm. All above error bars represent standard errors. d, Merged white-field/GFP/tdTomato images of day 4 exoplant cultures of backskin samples from the above Fig. 2j mice at P50 before Wax, and quantification (n = 6 exoplants separated from 3 mice) of GFP + % among tdTomato+ cells in the MK sheets. Scale bars: 100 μm. For all relevant figures, data are represented as mean ± SEM. P values were calculated with two tail t-test.
Extended Data Fig. 5 Conditional miR-31 ablation inhibits stress-induced HFSC transepidermal differentiation and premature aging.
a, Fluorescent images of backskin sections of P50 Tam-labeled Sox9-CreER;mTmG WT/cKO littermates at day 2 post Tam injection. Scale bars: 50μm. GFP is in green, tdTomato is in red. b, IF staining of backskin sections of P50 Tam-labeled Sox9-CreER;mTmG WT littermates at day 2 post Tam injection. Arrow head indicate c-Kit+ melonocytes. Scale bar: 50μm. c, Representative FACS sorting profiles of CD49f+GFP+(G+) and CD49f+GFP- (G-) cells from backskin epithelium of Tam-labeled Sox9-CreER;mTmG WT/cKO littermates at day 2 post Tam injection. d, DNA gel electrophoresis of genomic DNA PCR products of Mir31 surrounding regions. G-/G+: FACS sorted CD49f + GFP + and CD49f + GFP- cells described in B. Left labels: DNA size marker in bp, Right labels: expected PCR band positions and bp size of the indicated alleles. e, Upper panel: schematic of mouse genomic region around Mir31 gene and positions of the PCR primers used above (arrows). No other coding genes are present in vicinity of Mir31. Lower panel: DNA sequencing results of the indicated genomic region in the FACS sorted CD49f + GFP + backskin epithelium cells from Tam-labeled Sox9-CreER;mTmG WT/cKO littermates. f, Backskin photographs of WT/cKO littermates at indicated ages. The mice were Tam injected at P19. Scale bars: 1 cm. g, Backskin photographs of a pair of WT/cKO littermates at their day 0 (d0, P50), day 8 (d8) and day 10 (d10) post Waxing. The mice were Tam injected 2 days before waxing at P48. Scale bars, 1 cm. h, IF staining of the above g mice backskin sections at day 14 post wax. Scale bars: 50μm.
Extended Data Fig. 6 Conditional miR-31 ablation inhibits stress-induced HFSC transepidermal differentiation and premature aging.
a, Representative FACS profiles of CD49fhiCD34 + HFSCs from Rad/noRad skin regions of LIR treated WT/cKO littermates at A2 point. Box and % indicate CD49fhiCD34 + HFSCs and their relative abundance among CD49f + skin epithelia cells respectively. b, IF staining images of the LIR WT/cKO littermates at A2 analysis point. Scale bars: 50μm. Arrow heads: HFSC position. c, Backskin surface images and hair length quantification (n = 20 samples separated from 4 mice) of LIR WT/cKO littermates at days(d) post Wax2. Scale bars: 500μm. d, Quantification of white hair percentage in the above WT Rad and cKO-Rad skin regions at A2 point. n = 4 biologically independent mice. e, Quantification of relative GFP migration area (vs WT-noRad) in Fig. 3j. All above error bars represent standard errors. n = 6 exoplants separated from 3 mice. f, g, Comet analysis of epidermal cells (HF + IFE) from LIR WT/cKO littermates at different time points after 5-Gy LIR irradiation. f: Representative tail moments staining images, scale bar: 200 μm. g: Quantification of the tail moments, n = 30 cells for each. For all relevant figures, data are represented as mean ± SEM. P values were calculated with two tail t-test.
Extended Data Fig. 7 miR-31 promotes HFSCs transepidermal differentiation and aging by activating MAPK/ERK.
a, Representative FACS sorting profiles of CD49fhiCD34 + HFSCs (box) from P52 WT/DTG littermates (+Dox since P48 and waxed at P50 as indicated in the top diagram). % numbers indicate percentage of the HFSCs among the CD49f + epithelial cells. b, Volcano plot of gene expression changes between backskin HFSCs from WT/DTG littermates based on RNA-seq analysis. Each data point represents a gene. Red points indicate significantly changed genes (Padj < 0.01, Padj values were two-side and multiple comparison adjusted P values calculated by Deseq2). c, Western blots of cultured MK or NHEK cells stably expressing miR-31 or scrambled control miRNA (Scr) as indicated in Fig. 5b. Left labels: primary antibody used. Numbers indicate relative quantification (vs MK Scr for left, vs NHEK Scr for right). Green signals were first normalized to reference protein β-actin (ACT). d, Left: Western-blots of EDTA isolated backskin epithelium from young (8 weeks)/old (22 months) mice. Numbers indicate relative quantification of their above green bands. Right: Quantification of the left pERK bands. n = 3 replicated wells. e, Western blots of ERK/pERK in backskin epithelium of WT/DTG littermates treated with Trametinib (+T) or solvent control (+S). Numbers indicate relative quantification (vs WT + S). f, Western blots of ERK/pERK in Scr or miR-31 overexpressing MK cells treated with Trametinib (+T) or solvent control (+S). Numbers indicate relative quantification (vs Scr+S). g, Representative FACS profiles for backskin epithelium HFSC abundance quantification in Fig. 5g. h, Representative backskin photographs of Dox induced, waxed, and sch772984 (+sch) or solvent control (+S) treated WT/DTG littermates at d15 post waxing. Top diagram: treatment procedures. Scale bars=1 cm. For all relevant figures, data are represented as mean ± SEM. P values were calculated with two tail t-test.
Extended Data Fig. 8 Pharmaceutical inhibition of MAPK/ERK suppresses IR induced premature skin aging.
a, Left diagram: Experimental design for pharmaceutical intervention of LIR phenotypes using sch772984 (+sch) or Solvent control (+S) by Oral Gavage. Right panel: Backskin photographs of WT C57BL/6 mice at A2 analysis point that were LIR treated (LIR) or untreated (Ctrl), and were +sch or +S intervened. Scale bar: 1 cm. b,FACS profiles of HFSC abundance of +S/+ T LIR treated mice at A1 time. c, Comparative Pharmacological kinetics (PK) analysis of plasma drug concentration after one time same dosage Trametinib administration by Oral or Topical method. Horizon axis: hours post administration. n = 3 mice. d, Day 6 backskin exoplant cultures from A1 time backskins of mice in Fig. 6i and MK area quantification (n = 5 exoplants separated from 3 mice). Scale bars: 100 μm. e, SG size quantification in Fig. 6f. n = 5 HFs. f, Photos and quantification (n = 5) of backskin full-thickness wound healing of mice in Fig. 6i at A1 time. Scale bars: 5 mm. P values are for Rad+T vs Rad+S, n = 3 mice. g, FACS profiles and quantification (n = 3 mice) of HFSC abundance in Fig. 6i at A1 time. h, IF images of backskin sections of the Fig. 6j mice after +T or +S administration for 3 times without waxing and radiation. DAPI is in blue. Scale bars: 50μm. For all relevant figures, data are represented as mean ± SEM. P values were calculated with two tail t-test.
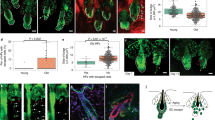
Extended Data Fig. 9 Clock is a key miR-31 target in regulating ERK activation and HFSC fate.
a, FACS sorting profiles of the CD49fhiCD34+ HFSCs (box) from backskin epithelium of P48 DTG and WT mice after 8hrs’ of Dox induction as indicated in the top diagram.b, Volcano plot of gene expression changes in RNA-seq datasets. Left: backskin HFSCs sorted from WT/DTG littermate 8 hours post Dox induction. Right: MK transfected by miR-31 mimic (Mim-31) or scramble mimic (Mim-Scr) (right) 8 hours post the transfection. Each data point represents a gene. Red points indicated significantly changed genes (Padj < 0.01, Padj values were two-side and multiple comparison adjusted P vaules calculated by Deseq2). c, GSEA analysis results of the b datasets as indicated using Targetscan predicted conserved miR-31 targets. Horizon axis: Log2 Fold of gene expression changes. Left side is positive. NES: normalized enrichment score; FDR: false discovery rate-adjusted P value of enrichment. d, Volcano plot of several reported miR-31 target genes expression changes in b RNA-seq datasets. HFSC-8hr: backskin HFSCs sorted from WT/DTG littermate 8 hours post Dox induction. MK-6hr: MK transfected by miR-31 mimic (Mim-31) or scramble mimic (Mim-Scr) (right) 8 hours post the transfection. e, GSEA analysis results using following Gene sets. Clock sig: genes that were enriched for >1.5 fold in Clock+ HFSCs vs Clock- HFSCs)35; KRAS sig: genes that were enriched for >1.5 fold in KrasG12D mutanxt HFSCs v.s. control littermate HFSCs30. Gene expression change datasets used: DTG/WT-4d-dataset: DTG/WT HFSCs in the 4d-dataset described in Fig. 5a; Clock+/Clock–: gene expression changes of Clock+ HFSCs vs Clock- HFSCs35.f, Diagram of predicted miR-31 binding sites on Clock 3’ UTR (upper panel) and its mutated version used for luciferase reporter assays (lower panel). g, Western blots of CLOCK expression in NHEK cells stably overexpressing miR-31 or scramble control (Scr) as indicated. h, Western blots of ERK/pERK and CLOCK in NHEKs stably expressing two different shRNAs against human CLOCK (shCLK1 and shCLK2) or scramble control shRNA (shScr). Numbers indicate relative quantification (vs shScr). i, Upper: Schematic of Teton-GFP transgenic skin graft generation by chamber transplantation. Middle: Photographs of transgenic skin grafts generated before and after Dox. Scale bars: 1 cm. Lower: Fluorescence microscope images of sections of Teton-GFP transgenic skin grafts with or without Dox induction as indicated in top diagram. GFP is in green. DAPI is in blue. Scale bars: 50μm. Dashed line indicates basement membrane. Scale bars, 50μm. j,qRT-PCR analysis of miRNA expressions in NHEK 24 hours after different dosages (0mJ/40mJ/60mJ) of one time UVB treatment. miR-31: miR-31-5p, miR-205: miR-205-5p. Vertical axis: relative expression % vs 0mJ. n = 3 biologically independent samples. k, Western blots of NHEKs after different dosages (0mJ/40mJ/60mJ) of one time UVB treatment. Numbers indicate relative quantification (vs 0mJ).For all relevant figures, data are represented as mean ± SEM. P values were calculated with two tail t-test.
Supplementary information
Source data
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Yu, Y., Zhang, X., Liu, F. et al. A stress-induced miR-31–CLOCK–ERK pathway is a key driver and therapeutic target for skin aging. Nat Aging 1, 795–809 (2021). https://doi.org/10.1038/s43587-021-00094-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-021-00094-8
This article is cited by
-
Cellular rejuvenation: molecular mechanisms and potential therapeutic interventions for diseases
Signal Transduction and Targeted Therapy (2023)
-
Biomarkers of aging
Science China Life Sciences (2023)
-
Single-cell transcriptomics reveals lineage trajectory of human scalp hair follicle and informs mechanisms of hair graying
Cell Discovery (2022)
-
Inhibition of microRNA turns back the CLOCK of hair follicle aging
Nature Aging (2021)