Abstract
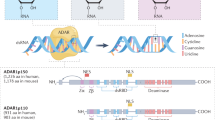
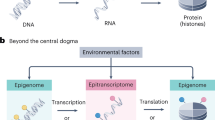
The mechanistic causes of aging, the time-related decline in function and good health that leads to increased mortality, remain poorly understood. Here we propose that age-dependent alteration of the epitranscriptome, encompassing more than 150 chemically distinct post-transcriptional modifications or editing events, warrants exploration as an important modulator of aging. The epitranscriptome is a potent regulator of RNA function, diverse cellular processes and tissue regenerative capacity. To date, only a few studies link alterations in the epitranscriptome to molecular and physiological changes during aging; however, epitranscriptome dysfunction is associated with and underlies several age-associated pathologies, including cancer and neurodegenerative, cardiovascular and autoimmune diseases. For example, changes in RNA modifications (such as N6-methyladenosine and inosine) impact cardiac physiology and are linked to cardiac fibrosis. Although an uncharted research focus, mapping epitranscriptome alterations in the context of aging may elucidate novel predictors of both health and lifespan, and may identify therapeutic targets for attenuating aging and abrogating age-related diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Niccoli, T. & Partridge, L. Ageing as a risk factor for disease. Curr. Biol. 22, R741–R752 (2012).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013). Landmark review article describing the hallmarks of aging—great introduction for newcomers to the aging field.
Wilkinson, G. S. & South, J. M. Life history, ecology and longevity in bats. Aging Cell 1, 124–131 (2002).
Buffenstein, R. et al. Probing pedomorphy and prolonged lifespan in naked mole-rats and dwarf mice. Physiology 35, 96–111 (2020).
Benayoun, B. A., Pollina, E. A. & Brunet, A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat. Rev. Mol. Cell Biol. 16, 593–610 (2015).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Gilbert, W. V., Bell, T. A. & Schaening, C. Messenger RNA modifications: form, distribution, and function. Science 352, 1408–1412 (2016).
McCown, P. J. et al. Naturally occurring modified ribonucleosides. Wiley Interdiscip. Rev. RNA 11, e1595 (2020). Comprehensive review article on RNA modifications—great introduction for newcomers to the RNA modification field.
Wiener, D. & Schwartz, S. The epitranscriptome beyond m6A. Nat. Rev. Genet. 22, 119–131 (2021).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Squires, J. E. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033 (2012).
Zhang, L. S. et al. Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol. Cell 74, 1304–1316 (2019).
Dai, Q. et al. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods 14, 695–698 (2017).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014). In the manuscript, pseudouridine modifications were profiled transcriptome-wide and found on human mRNA and ncRNAs.
Bazak, L. et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24, 365–376 (2014).
Littlefield, J. W. & Dunn, D. B. Natural occurrence of thymine and three methylated adenine bases in several ribonucleic acids. Nature 181, 254–255 (1958).
Davis, F. F. & Allen, F. W. Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem. 227, 907–915 (1957).
Helm, M. & Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18, 275–291 (2017).
Su, D. et al. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc. 9, 828–841 (2014).
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018). This article describes a database containing all known RNA modification pathways.
Nachtergaele, S. & He, C. Chemical modifications in the life of an mRNA transcript. Annu. Rev. Genet. 52, 349–372 (2018).
Shulman, Z. & Stern-Ginossar, N. The RNA modification N6-methyladenosine as a novel regulator of the immune system. Nat. Immunol. 21, 501–512 (2020).
Jonkhout, N. et al. The RNA modification landscape in human disease. RNA 23, 1754–1769 (2017).
Stojkovic, V. & Fujimori, D. G. Mutations in RNA methylating enzymes in disease. Curr. Opin. Chem. Biol. 41, 20–27 (2017).
Barbieri, I. & Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 20, 303–322 (2020).
Abe, M. et al. Impact of age-associated increase in 2′-O-methylation of miRNAs on aging and neurodegeneration in Drosophila. Genes Dev. 28, 44–57 (2014).
Min, K. W. et al. Profiling of m6A RNA modifications identified an age-associated regulation of AGO2 mRNA stability. Aging Cell 17, e12753 (2018). This paper maps and quantifies m6A modifications in peripheral blood mononuclear cells from young and old individuals and reveals a global decrease in m6A RNA modifications with increasing age.
Nicholas, A. et al. Age-related gene-specific changes of A-to-I mRNA editing in the human brain. Mech. Ageing Dev. 131, 445–447 (2010).
Matera, A. G. & Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15, 108–121 (2014).
Dahlberg, A. E. The functional role of ribosomal RNA in protein synthesis. Cell 57, 525–529 (1989).
Kirchner, S. & Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 16, 98–112 (2015).
Fabian, M. R., Sonenberg, N. & Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379 (2010).
Kim, N. K., Theimer, C. A., Mitchell, J. R., Collins, K. & Feigon, J. Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res. 38, 6746–6756 (2010).
Tang, H. et al. HuR regulates telomerase activity through TERC methylation. Nat. Commun. 9, 2213 (2018). In the manuscript, the authors identify m5C modifications in TERC and demonstrate that m5C modifications impact telomerase activity.
Zaccara, S., Ries, R. J. & Jaffrey, S. R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624 (2019).
Wilusz, C. J., Wormington, M. & Peltz, S. W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2, 237–246 (2001).
Wang, X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014).
Sommer, S., Lavi, U. & Darnell, J. E. Jr The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J. Mol. Biol. 124, 487–499 (1978).
Geula, S. et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015).
Zhang, C. et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276 (2017).
Zaccara, S. & Jaffrey, S. R. A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell https://doi.org/10.1016/j.cell.2020.05.012 (2020).
Lasman, L. et al. Context-dependent functional compensation between Ythdf m6A reader proteins. Genes Dev. 34, 1373–1391 (2020).
Du, H. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 7, 12626 (2016).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Huang, H. et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018).
Tuorto, F. et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19, 900–905 (2012).
Blanco, S. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 33, 2020–2039 (2014). In the study, the authors demonstrate a role for NSUN2-mediated m5C tRNA modifications and tRNA-derived fragments in modulating cellular response to stress and in neurodevelopment.
Lee, B. P., Smith, M., Buffenstein, R. & Harries, L. W. Negligible senescence in naked mole rats may be a consequence of well-maintained splicing regulation. Geroscience 42, 633–651 (2020).
Deschenes, M. & Chabot, B. The emerging role of alternative splicing in senescence and aging. Aging Cell 16, 918–933 (2017).
Karijolich, J. & Yu, Y. T. Spliceosomal snRNA modifications and their function. RNA Biol. 7, 192–204 (2010).
Hasler, D. et al. The Alazami syndrome-associated protein LARP7 guides U6 small nuclear RNA modification and contributes to splicing robustness. Mol. Cell 77, 1014–1031 (2020).
Wang, X. et al. LARP7-mediated U6 snRNA modification ensures splicing fidelity and spermatogenesis in mice. Mol. Cell 77, 999–1013 (2020).
Rueter, S. M., Dawson, T. R. & Emeson, R. B. Regulation of alternative splicing by RNA editing. Nature 399, 75–80 (1999).
Solomon, O. et al. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR). RNA 19, 591–604 (2013).
Zhao, X. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24, 1403–1419 (2014).
Liu, N. et al. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518, 560–564 (2015).
Sloan, K. E. et al. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14, 1138–1152 (2017).
Marcel, V. et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 24, 318–330 (2013).
Yoon, A. et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 312, 902–906 (2006).
Nachmani, D. et al. Germline NPM1 mutations lead to altered rRNA 2′-O-methylation and cause dyskeratosis congenita. Nat. Genet. 51, 1518–1529 (2019). This study describes how novel mutations in NPM1 cause dyskeratosis congenita and decrease site-specific ribosomal RNA Nm modifications.
Taoka, M. et al. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 46, 9289–9298 (2018).
Ignatova, V. V. et al. The rRNA m6A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 34, 715–729 (2020).
Begley, U. et al. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell 28, 860–870 (2007).
Laxman, S. et al. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell 154, 416–429 (2013).
Nedialkova, D. D. & Leidel, S. A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–1618 (2015).
Rapino, F. et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 558, 605–609 (2018). In this manuscript, inhibition of RNA-modifying enzymes involved in tRNA U34 modification restores sensitivity to BRAF inhibition in drug-resistant melanomas.
Thiaville, P. C. et al. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell 3, 29–45 (2016).
Pollo-Oliveira, L. et al. Loss of elongator- and KEOPS-dependent tRNA modifications leads to severe growth phenotypes and protein aggregation in yeast. Biomolecules https://doi.org/10.3390/biom10020322 (2020).
Chan, C. T. et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 3, 937 (2012).
Fu, D. et al. Human AlkB homolog ABH8 is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 30, 2449–2459 (2010).
Endres, L. et al. Alkbh8 regulates selenocysteine-protein expression to protect against reactive oxygen species damage. PLoS ONE 10, e0131335 (2015).
Wang, X. et al. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Choe, J. et al. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560 (2018).
Mao, Y. et al. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 10, 5332 (2019).
Zhou, J. et al. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015).
Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96 (2016).
North, B. J. & Sinclair, D. A. The intersection between aging and cardiovascular disease. Circ. Res. 110, 1097–1108 (2012).
Mathiyalagan, P. et al. FTO-dependent N6-methyladenosine regulates cardiac function during remodeling and repair. Circulation 139, 518–532 (2019).
Kmietczyk, V. et al. m6A-mRNA methylation regulates cardiac gene expression and cellular growth. Life Sci. Alliance https://doi.org/10.26508/lsa.201800233 (2019).
Dorn, L. E. et al. The N6-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation 139, 533–545 (2019).
Frayling, T. M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
Liu, C., Mou, S. & Pan, C. The FTO gene rs9939609 polymorphism predicts risk of cardiovascular disease: a systematic review and meta-analysis. PLoS ONE 8, e71901 (2013).
Jia, G. et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Mauer, J. et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017).
Wei, J. et al. Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71, 973–985 (2018).
Moore, J. B. T. et al. The A-to-I RNA editing enzyme Adar1 is essential for normal embryonic cardiac growth and development. Circ. Res. 127, 550–552 (2020).
El Azzouzi, H. et al. Cardiomyocyte specific deletion of ADAR1 causes severe cardiac dysfunction and increased lethality. Front. Cardiovasc. Med. 7, 30 (2020).
Fei, J., Cui, X. B., Wang, J. N., Dong, K. & Chen, S. Y. ADAR1-mediated RNA editing, a novel mechanism controlling phenotypic modulation of vascular smooth muscle cells. Circ. Res. 119, 463–469 (2016).
Stellos, K. et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 22, 1140–1150 (2016). This article implicates elevated A-to-I editing of the mRNA encoding cathepsin S in the pathogenesis of atherosclerosis.
Suzuki, T., Nagao, A. & Suzuki, T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 45, 299–329 (2011).
Fulop, T. et al. Immunosenescence and inflamm-aging as two sides of the same coin: friends or foes? Front. Immunol. 8, 1960 (2017).
O’Connell, M. A., Mannion, N. M. & Keegan, L. P. The epitranscriptome and innate immunity. PLoS Genet. 11, e1005687 (2015).
Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005). Key study demonstrating that delivery of modified RNA suppresses RNA recognition by Toll-like receptors.
Nelson, J. et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 6, eaaz6893 (2020).
Lu, M. et al. N6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 5, 584–598 (2020).
Agrawal, A. Mechanisms and implications of age-associated impaired innate interferon secretion by dendritic cells: a mini-review. Gerontology 59, 421–426 (2013).
Hartner, J. C., Walkley, C. R., Lu, J. & Orkin, S. H. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 10, 109–115 (2009).
George, C. X., Ramaswami, G., Li, J. B. & Samuel, C. E. Editing of cellular self-RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J. Biol. Chem. 291, 6158–6168 (2016).
Rice, G. I. et al. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat. Genet. 44, 1243–1248 (2012).
Liu, E. Y., Cali, C. P. & Lee, E. B. RNA metabolism in neurodegenerative disease. Dis. Models Mech. 10, 509–518 (2017).
Reitz, C. et al. Genetic variants in the fat and obesity associated (FTO) gene and risk of Alzheimer’s disease. PLoS ONE 7, e50354 (2012).
Yamashita, T. et al. Rescue of amyotrophic lateral sclerosis phenotype in a mouse model by intravenous AAV9-ADAR2 delivery to motor neurons. EMBO Mol. Med. 5, 1710–1719 (2013).
Hideyama, T. et al. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol. Dis. 45, 1121–1128 (2012).
Brusa, R. et al. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 270, 1677–1680 (1995).
Higuchi, M. et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81 (2000).
Yamashita, T. et al. A role for calpain-dependent cleavage of TDP-43 in amyotrophic lateral sclerosis pathology. Nat. Commun. 3, 1307 (2012).
Moore, S. et al. ADAR2 mislocalization and widespread RNA editing aberrations in C9orf72-mediated ALS/FTD. Acta Neuropathol. 138, 49–65 (2019).
Parikshak, N. N., Gandal, M. J. & Geschwind, D. H. Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders. Nat. Rev. Genet. 16, 441–458 (2015).
Flores, J. V. et al. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Rep. 8, 112–124 (2017).
Braun, D. A. et al. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet. 49, 1529–1538 (2017).
Arrondel, C. et al. Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat. Commun. 10, 3967 (2019).
Livneh, I., Moshitch-Moshkovitz, S., Amariglio, N., Rechavi, G. & Dominissini, D. The m6A epitranscriptome: transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 21, 36–51 (2020).
White, M. C. et al. Age and cancer risk: a potentially modifiable relationship. Am. J. Prev. Med. 46, S7–S15 (2014).
Shen, S. et al. An epitranscriptomic mechanism underlies selective mRNA translation remodelling in melanoma persister cells. Nat. Commun. 10, 5713 (2019).
Vu, L. P. et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376 (2017). This and the study below identify a role for m6A RNA modifications in normal hematopoiesis and in promoting leukemia.
Weng, H. et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell 22, 191–205 (2018). This and the study above identify a role for m6A RNA modifications in normal hematopoiesis and in promoting leukemia.
Han, D. et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 566, 270–274 (2019).
Ishizuka, J. J. et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 565, 43–48 (2019).
Ruggero, D. et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299, 259–262 (2003).
Alter, B. P., Giri, N., Savage, S. A. & Rosenberg, P. S. Cancer in dyskeratosis congenita. Blood 113, 6549–6557 (2009).
Ronchetti, D. et al. The expression pattern of small nucleolar and small Cajal body-specific RNAs characterizes distinct molecular subtypes of multiple myeloma. Blood Cancer J. 2, e96 (2012).
Valleron, W. et al. Small nucleolar RNA expression profiling identifies potential prognostic markers in peripheral T-cell lymphoma. Blood 120, 3997–4005 (2012).
McMahon, M. et al. A single H/ACA small nucleolar RNA mediates tumor suppression downstream of oncogenic RAS. eLife https://doi.org/10.7554/eLife.48847 (2019).
Babaian, A. et al. Loss of m1acp3Ψ ribosomal RNA modification is a major feature of cancer. Cell Rep. 31, 107611 (2020).
Lee, M. Y., Leonardi, A., Begley, T. J. & Melendez, J. A. Loss of epitranscriptomic control of selenocysteine utilization engages senescence and mitochondrial reprogramming. Redox Biol. 28, 101375 (2020).
Li, Q. et al. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J. Cell. Biochem. 118, 2587–2598 (2017).
Bellodi, C., Kopmar, N. & Ruggero, D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 29, 1865–1876 (2010).
Wang, Y. et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21, 195–206 (2018).
Xiang, Y. et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576 (2017).
Zhang, C. et al. METTL3 and N6-methyladenosine promote homologous recombination-mediated repair of DSBs by modulating DNA–RNA hybrid accumulation. Mol. Cell 79, 425–442 (2020).
De Jesus, D. F. et al. m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes. Nat. Metab. 1, 765–774 (2019).
Fakruddin, M. et al. Defective mitochondrial tRNA taurine modification activates global proteostress and leads to mitochondrial disease. Cell Rep. 22, 482–496 (2018).
Lin, H. et al. CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat. Commun. 9, 1875 (2018).
Pang, W. W. et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl Acad. Sci. USA 108, 20012–20017 (2011).
Mejia-Ramirez, E. & Florian, M. C. Understanding intrinsic hematopoietic stem cell aging. Haematologica 105, 22–37 (2020).
Li, Z. et al. Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 28, 904–917 (2018).
Guzzi, N. et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216 (2018).
Gu, B. W., Fan, J. M., Bessler, M. & Mason, P. J. Accelerated hematopoietic stem cell aging in a mouse model of dyskeratosis congenita responds to antioxidant treatment. Aging Cell 10, 338–348 (2011).
Kirwan, M. & Dokal, I. Dyskeratosis congenita: a genetic disorder of many faces. Clin. Genet. 73, 103–112 (2008).
Verdu, E., Ceballos, D., Vilches, J. J. & Navarro, X. Influence of aging on peripheral nerve function and regeneration. J. Peripher. Nerv. Syst. 5, 191–208 (2000).
Weng, Y. L. et al. Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97, 313–325 (2018).
Murtha, L. A. et al. The role of pathological aging in cardiac and pulmonary fibrosis. Aging Dis. 10, 419–428 (2019).
Kropski, J. A. et al. A novel dyskerin (DKC1) mutation is associated with familial interstitial pneumonia. Chest 146, e1–e7 (2014).
Ben-Shoshan, S. O. et al. ADAR1 deletion induces NFκB and interferon signaling dependent liver inflammation and fibrosis. RNA Biol. 14, 587–602 (2017). In this manuscript it was shown that ADAR1 depletion in vivo induces inflammation and liver fibrosis.
Hebras, J., Krogh, N., Marty, V., Nielsen, H. & Cavaille, J. Developmental changes of rRNA ribose methylations in the mouse. RNA Biol. 17, 150–164 (2020).
Shafik, A. M. et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 22, 17 (2021).
Asadi Shahmirzadi, A. et al. Alpha-ketoglutarate, an endogenous metabolite, extends lifespan and compresses morbidity in aging mice. Cell Metab. 32, 447–456 (2020).
Thomas, J. M., Batista, P. J. & Meier, J. L. Metabolic regulation of the epitranscriptome. ACS Chem. Biol. 14, 316–324 (2019).
Fedeles, B. I., Singh, V., Delaney, J. C., Li, D. & Essigmann, J. M. The AlkB family of Fe(II)/alpha-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 290, 20734–20742 (2015).
Wu, Z. et al. METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 48, 11083–11096 (2020). In this study, it was found that m6A is decreased in stem cell models of the progeroid syndromes Hutchinson–Gilford progeria syndrome and Werner syndrome.
Sebastiani, P. et al. RNA editing genes associated with extreme old age in humans and with lifespan in C. elegans. PLoS ONE 4, e8210 (2009). This article and the one below describe SNPs in factors involved in A-to-I editing and m6A modifications and their association with human longevity.
Cardelli, M. et al. A polymorphism of the YTHDF2 gene (1p35) located in an Alu-rich genomic domain is associated with human longevity. J. Gerontol. A 61, 547–556 (2006). This article and the one above describe SNPs in factors involved in A-to-I editing and m6A modifications and their association with human longevity.
Schosserer, M. et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 6, 6158 (2015).
Liberman, N. et al. N6-adenosine methylation of ribosomal RNA affects lipid oxidation and stress resistance. Sci. Adv. 6, eaaz4370 (2020).
Heissenberger, C. et al. Loss of the ribosomal RNA methyltransferase NSUN5 impairs global protein synthesis and normal growth. Nucleic Acids Res. 47, 11807–11825 (2019).
Khoddami, V. et al. Transcriptome-wide profiling of multiple RNA modifications simultaneously at single-base resolution. Proc. Natl Acad. Sci. USA 116, 6784–6789 (2019).
Marchand, V. et al. AlkAniline-Seq: profiling of m7G and m3C RNA modifications at single nucleotide resolution. Angew. Chem. Int. Ed. 57, 16785–16790 (2018).
Liu, H. et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat. Commun. 10, 4079 (2019).
Heiss, M., Reichle, V. F. & Kellner, S. Observing the fate of tRNA and its modifications by nucleic acid isotope labeling mass spectrometry: NAIL-MS. RNA Biol. 14, 1260–1268 (2017).
Liu, X. M., Zhou, J., Mao, Y., Ji, Q. & Qian, S. B. Programmable RNA N6-methyladenosine editing by CRISPR-Cas9 conjugates. Nat. Chem. Biol. 15, 865–871 (2019).
Richard, E. M. et al. Bi-allelic variants in METTL5 cause autosomal-recessive intellectual disability and microcephaly. Am. J. Hum. Genet. 105, 869–878 (2019).
Braun, D. A. et al. Mutations in WDR4 as a new cause of Galloway–Mowat syndrome. Am. J. Med. Genet. A 176, 2460–2465 (2018).
Monies, D., Vagbo, C. B., Al-Owain, M., Alhomaidi, S. & Alkuraya, F. S. Recessive truncating mutations in ALKBH8 cause intellectual disability and severe impairment of wobble uridine modification. Am. J. Hum. Genet. 104, 1202–1209 (2019).
de Brouwer, A. P. M. et al. Variants in PUS7 cause intellectual disability with speech delay, microcephaly, short stature, and aggressive behavior. Am. J. Hum. Genet. 103, 1045–1052 (2018).
de Paiva, A. R. B. et al. PUS3 mutations are associated with intellectual disability, leukoencephalopathy, and nephropathy. Neurol. Genet. 5, e306 (2019).
Shen, F. et al. Decreased N6-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J. Clin. Endocrinol. Metab. 100, E148–E154 (2015).
Vlachogiannis, N. I. et al. Increased adenosine-to-inosine RNA editing in rheumatoid arthritis. J. Autoimmun. 106, 102329 (2020).
Delaunay, S. et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J. Exp. Med. 213, 2503–2523 (2016).
Ghezzi, D. et al. Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am. J. Hum. Genet. 90, 1079–1087 (2012).
Kopajtich, R. et al. Mutations in GTPBP3 cause a mitochondrial translation defect associated with hypertrophic cardiomyopathy, lactic acidosis, and encephalopathy. Am. J. Hum. Genet. 95, 708–720 (2014).
Zhang, X. et al. Small RNA modifications in Alzheimer’s disease. Neurobiol. Dis. 145, 105058 (2020).
Liddicoat, B. J. et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120 (2015).
Acknowledgements
We thank J. Kimmel and R. Cohen at Calico Life Sciences LLC as well as D. Fujimori (UCSF) and C. Stumpf (eFFECTOR Therapeutics) for their helpful comments on this article. Figures were created using BioRender.com. M.M. and R.B. have been funded by Calico Life Sciences LLC. C.F. was funded by the NIH/NIDDK (1K08DK119561-01) and the American Society of Hematology (A132428).
Author information
Authors and Affiliations
Contributions
M.M. and R.B. led article conceptualization and writing. M.M., C.F. and R.B. researched data for the article. M.M., C.F. and R.B. discussed the content and contributed to writing and editing of the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
M.M. and R.B. are employees of Calico Life Sciences LLC. C.F. declares no competing interests.
Additional information
Peer review information Nature Aging thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
List of abbreviations used.
Rights and permissions
About this article
Cite this article
McMahon, M., Forester, C. & Buffenstein, R. Aging through an epitranscriptomic lens. Nat Aging 1, 335–346 (2021). https://doi.org/10.1038/s43587-021-00058-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-021-00058-y
This article is cited by
-
Molecular hallmarks of ageing in amyotrophic lateral sclerosis
Cellular and Molecular Life Sciences (2024)
-
m6A epitranscriptomic regulation of tissue homeostasis during primate aging
Nature Aging (2023)