Abstract
Hair follicles, mammalian mini-organs that grow hair, miniaturize during aging, leading to hair thinning and loss. Here we report that hair follicle stem cells (HFSCs) lose their regenerative capabilities during aging owing to the adoption of an atypical cell division program. Cell fate tracing and cell division axis analyses revealed that while HFSCs in young mice undergo typical symmetric and asymmetric cell divisions to regenerate hair follicles, upon aging or stress, they adopt an atypical ‘stress-responsive’ type of asymmetric cell division. This type of division is accompanied by the destabilization of hemidesmosomal protein COL17A1 and cell polarity protein aPKCλ and generates terminally differentiating epidermal cells instead of regenerating the hair follicle niche. With the repetition of these atypical divisions, HFSCs detach from the basal membrane causing their exhaustion, elimination and organ aging. The experimentally induced stabilization of COL17A1 rescued organ homeostasis through aPKCλ stabilization. These results demonstrate that distinct stem cell division programs may govern tissue and organ aging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request. Microarray data were deposited in Gene Expression Omnibus (www.ncbi.nlm.gov/geo/) under series identifier GSE124119.
References
Signer, R. A. & Morrison, S. J. Mechanisms that regulate stem cell aging and life span. Cell Stem Cell 12, 152–165 (2013).
Matsumura, H. et al. Hair follicle aging is driven by transepidermal elimination of stem cells via COL17A1 proteolysis. Science 351, aad4395 (2016).
Liu, N. et al. Stem cell competition orchestrates skin homeostasis and ageing. Nature 568, 344–350 (2019).
Nishimura, E. K., Granter, S. R. & Fisher, D. E. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science 307, 720–724 (2005).
Baar, M. P. et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 169, 132–147 (2017).
Baker, D. J. et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature 530, 184–189 (2016).
van Deursen, J. M. The role of senescent cells in ageing. Nature 509, 439–446 (2014).
Poulson, N. D. & Lechler, T. Asymmetric cell divisions in the epidermis. Int. Rev. Cell Mol. Biol. 295, 199–232 (2012).
Morrison, S. J. & Kimble, J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 441, 1068–1074 (2006).
Lechler, T. & Fuchs, E. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275–280 (2005).
Ray, S. & Lechler, T. Regulation of asymmetric cell division in the epidermis. Cell Div. 6, 12 (2011).
Goodell, M. A. & Rando, T. A. Stem cells and healthy aging. Science 350, 1199–1204 (2015).
Cheng, J. et al. Centrosome misorientation reduces stem cell division during ageing. Nature 456, 599–604 (2008).
Bast, L. et al. Increasing neural stem cell division asymmetry and quiescence are predicted to contribute to the age-related decline in neurogenesis. Cell Rep. 25, 3231–3240 (2018).
Florian, M. C. et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell 10, 520–530 (2012).
Florian, M. C. et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature 503, 392–396 (2013).
Goulas, S., Conder, R. & Knoblich, J. A. The par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell 11, 529–540 (2012).
Zovein, A. C. et al. Beta1 integrin establishes endothelial cell polarity and arteriolar lumen formation via a Par3-dependent mechanism. Dev. Cell 18, 39–51 (2010).
Durgan, J., Kaji, N., Jin, D. & Hall, A. Par6B and atypical PKC regulate mitotic spindle orientation during epithelial morphogenesis. J. Biol. Chem. 286, 12461–12474 (2011).
Hao, Y. et al. Par3 controls epithelial spindle orientation by aPKC-mediated phosphorylation of apical Pins. Curr. Biol. 20, 1809–1818 (2010).
Niessen, M. T. et al. aPKCλ controls epidermal homeostasis and stem cell fate through regulation of division orientation. J. Cell Biol. 202, 887–900 (2013).
Osada, S. et al. Atypical protein kinase c isoform, aPKCλ, is essential for maintaining hair follicle stem cell quiescence. J. Invest. Dermatol. 135, 2584–2592 (2015).
Greco, V. et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 4, 155–169 (2009).
Rompolas, P., Mesa, K. R. & Greco, V. Spatial organization within a niche as a determinant of stem-cell fate. Nature 502, 513–518 (2013).
Margadant, C., Charafeddine, R. A. & Sonnenberg, A. Unique and redundant functions of integrins in the epidermis. FASEB J. 24, 4133–4152 (2010).
Fine, J. D. et al. The classification of inherited epidermolysis bullosa (EB): report of the third international consensus meeting on diagnosis and classification of EB. J. Am. Acad. Dermatol. 58, 931–950 (2008).
Tanimura, S. et al. Hair follicle stem cells provide a functional niche for melanocyte stem cells. Cell Stem Cell 8, 177–187 (2011).
Hsu, Y. C., Pasolli, H. A. & Fuchs, E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144, 92–105 (2011).
Ito, M., Kizawa, K., Hamada, K. & Cotsarelis, G. Hair follicle stem cells in the lower bulge form the secondary germ, a biochemically distinct but functionally equivalent progenitor cell population, at the termination of catagen. Differentiation 72, 548–557 (2004).
Lough, K. J. et al. Telophase correction refines division orientation in stratified epithelia. eLife https://doi.org/10.7554/eLife.49249 (2019).
Lu, M. S. & Johnston, C. A. Molecular pathways regulating mitotic spindle orientation in animal cells. Development 140, 1843–1856 (2013).
Morin, X. & Bellaiche, Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell 21, 102–119 (2011).
Smith, C. A. et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant NUMB. EMBO J. 26, 468–480 (2007).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Toyoshima, F. & Nishida, E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 26, 1487–1498 (2007).
Suzuki, A. et al. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 14, 1425–1435 (2004).
Lay, K., Kume, T. & Fuchs, E. FOXC1 maintains the hair follicle stem cell niche and governs stem cell quiescence to preserve long-term tissue-regenerating potential. Proc. Natl Acad. Sci. USA 113, E1506–E1515 (2016).
He, S. & Sharpless, N. E. Senescence in health and disease. Cell 169, 1000–1011 (2017).
Inomata, K. et al. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell 137, 1088–1099 (2009).
Wang, J. et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 148, 1001–1014 (2012).
Morris, R. J. et al. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22, 411–417 (2004).
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71 (1999).
Kawamoto, S. et al. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 470, 263–268 (2000).
Olasz, E. B. et al. Human bullous pemphigoid antigen 2 transgenic skin elicits specific IgG in wild-type mice. J. Invest. Dermatol. 127, 2807–2817 (2007).
Niculescu, C. et al. Conditional ablation of integrin α-6 in mouse epidermis leads to skin fragility and inflammation. Eur. J. Cell Biol. 90, 270–277 (2011).
Hirose, T. et al. An essential role of the universal polarity protein, aPKCλ, on the maintenance of podocyte slit diaphragms. PLoS ONE 4, e4194 (2009).
de Boer, J. et al. A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol. Cell 1, 981–990 (1998).
Acknowledgements
We thank Y. Nishimori, E. Suzuki, A. Tsuda, M. Natsuki, T. Kato, R. Yajima, F. Toki, H. Shimizu, W. Nishie, K. Yancey, F. Matsuzaki, H. Nishina, F. Ishino, T. Usami and M. Kanai for their helpful support and DASS Manuscript for editing. E. Georges-Labouesse provided the source of the ITGA6 flox mouse line. E.K.N. is supported by AMED under grant no. JP17gm5010002–JP20gm5010002 and a Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research S (26221303). H.M. is supported by a Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research on Innovative Areas ‘Cell Competition’ (26114001), a Grant-in-Aid for Scientific Research B (18H02637), a Young Scientific Research B (26860258, 24790376) and a GSK Japan Research Grant (2017-B9).
Author information
Authors and Affiliations
Contributions
H. Matsumura and E.K.N. conceived the study and wrote the manuscript. H. Matsumura performed the majority of the experiments and analyzed the data. N.L., A.T. and Y.M. prepared aged mice. S.I. performed all TEM experiments. D.N. advised on in vitro keratinocyte analysis. S.K. provided human scalp specimens. H. Morinaga performed H2O2 experiments. A.D.A. provided Itga6 flox mice. S.O. provided aPKCλ flox mice.
Corresponding authors
Ethics declarations
Competing interests
E.K.N. is an advisor to EADERM Co., Ltd. All other authors have no competing interests.
Additional information
Peer review information Nature Aging thanks Claire Higgins, Lenhard Rudolph and Scott Williams for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
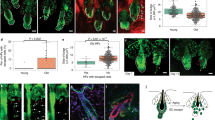
Extended Data Fig. 1 Asymmetric cell divisions from embryo to adult life.
a, Schematic representation of cell divisions. Symmetric cell divisions (SCDs) of stem cells generate two identical cells by orienting the mitotic spindle parallel to the underlying basement membrane. In contrast, the asymmetric cells divisions (ACDs) of stem cells generate a self-renewing stem cell and a differentiating cell by orienting the mitotic spindle perpendicular to the underlying basement membrane. By analyzing the cell division axes using phospho-histone H3 (PH3) with γ-tubulin (γ—TUB) (red asterisk, metaphase) or Survivin with DAPI (orange asterisk, anaphase), SCDs or ACDs are defined as cell division angle 0–30o or 60–90o against basement membrane, respectively. b, Schematic representation of spindle orientation in E16.5 embryonic basal cells (n = 21 cells). E16.5 embryonic basal cells undergo perpendicular cell division or parallel cell division as previously described (Lechter & Fuchs, 2005, Nature 437, 275–280; Williams et al., 2014, Nature Cell Biology, 16 758–769). c, Representative IF images of PH3, γ—TUB and PARD3 in E16.5 embryonic basal cells. Perpendicularly dividing cells against the basement membrane show polarized PARD3 expression at the apical side (arrowheads) (assumed as asymmetric cell division), while parallelly dividing cells against the basement-membrane show non-polarized PARD3 expression (assumed as symmetric cell division). d, Schematic representation of spindle orientation in bulge (n = 68 cells) and hair germ basal cells (n = 43 cells) and HFs with several stem cell markers at postnatal day 23 (p23). HFs at early anagen stage consist of four different types with different stem cell markers. The bulge and hair germ basal cells at p23 undergo perpendicular cell division or parallel cell division. e, f, Representative IF images of PH3, γ—TUB and PARD3 (e) or NUMB (f) in bulge basal cells (Bg) and in hair germ cells (Hg) at p23. In bulge basal cells, perpendicularly dividing cells showed polarized PARD3 expression at the apical side (arrowheads) (assumed as asymmetric cell division), while parallelly dividing cells showed non-polarized PARD3 expression (assumed as symmetric cell division). However, in hair germ cells, perpendicularly dividing cells do not show polarized PARD3 expression at the apical side. g, Quantification of percent cells with polarized localization of NUMB and PARD3 in Bg and Hg areas (Bg, n = 52; Hg, n = 32 cells). h, Quantification of percent cells with polarized expression of NUMB and PARD3 in Bg, sBg and bulb areas (Bg, n = 18; sBg, n = 55; Bulb, n = 6 cells). In Bg and sBg basal cells, cells dividing at a perpendicular angle (60–90°) significantly showed polarized expression (g, h). i, IF images of ITGA6, COL17A1 and LHX2 in HFs at p25. At this stage, old bulge and hair germ cells proliferate and generate new bulge cells. j, IF images of ITGA6, KRT6 and KRT75 in HFs from p25 to p28. k, Representative IF images of GFP, COL17A1 and Survivin at p23 from GFP-labeled HFSCs at p21. Perpendicularly dividing Survivin+ GFP cells generate COL17A1+ cells. l, Schematic representation of spindle orientation in bulge (n = 38) and sub-bulge (n = 67 cells) HFs with stem cell markers at 3 days after HC induction by depilation at 7–9 wo (n = 4 mice). m, Quantification of cell division angles in bulge basal cells at p23 or p25 or 3 days after HC induction at 7–9 wo. Bulge basal cells at 3 days after HC induction showed a similar cell division pattern to p25 bulge basal cells. n, 3D reconstructed IF images of Survivin in GFP labeled-HFSCs after HC induction. Perpendicular cell division patterns are subdivided as inner cell division and surface cell division. o, The frequency of type of cell division (n = 17 HFs) in (n). p, 3D reconstructed IF images of KRT75 and Survivin in HFSCs after HC induction. HFSCs occasionally perpendicularly divided and differentiated into KRT75+ cells at 3 days after HC induction (n, o, p). q, Schematic representation of hair regeneration after depilation at 8–11 wo. r, Representative IF images of PH3 in the bulge and hair germ areas at 3 days after HC induction. s, Quantification of the number of PH3+ mitotic cells in the bulge areas at 3 days after HC induction (8 wo, n = 30 HFs; 26 mo, n = 60 HFs). PH3+ mitotic cells in the bulge were significantly decreased by aging. t, Quantification of the postulated number of cell division angles in the bulge and sub-bulge areas at 3 days after HC induction. Data were calculated by the number of actual mitotic cells (Extended Data Fig. 1s) and the ratio of cell division angles (Fig. 1h). u, IF images of NUMB in HFSCs from young (8w) and from aged (26 mo) mice at 3 days after depilation. Arrowheads; polarized numb expression. v, Quantification of percent cells with polarized expression in Bg (young (7–12 wo), n = 9 mice, n = 35 cells; aged (24–27 mo), n = 3 mice, n = 17 cells) and in sBg (young (7–12 wo), n = 9 mice, n = 14 cells; aged (24–27 mo), n = 3 mice, n = 15 cells) areas. Aged HFSCs significantly lost their polarized expression. w, 3D reconstructed IF images of GFP, KRT10 and KRT75 in GFP labeled-HFSCs from young (11 wo) and from aged (25 mo) mice at 3 days after depilation. x, y, Representative IF images (x) and 3D reconstructed IF images (y) of GFP and KRT10 without hair depilation in GFP labeled-HFSCs from young and from aged mice. Frequency of KRT10+ HFs in the GFP+ HFSCs are shown at the bottom (n = 8–10 images in each group). Arrows, normal HFSCs; arrowheads; aged epidermally differentiated HFSCs. White line and fan-shaped line and upper number, spindle angle against basal cell layer (o) ; white dashed line, basal cell layer. Error bars, means ± SEM; Chi-square test (g, h, t, v) or two-tailed Mann-Whitney U-test (s).
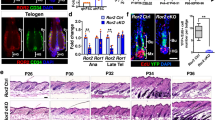
Extended Data Fig. 2 Phenotypic consequence of an HFSC specific deficiency of Col17a1 or Itga6 gene.
a, Representative IF images of COL17A1, Survivin and KRT10 in the bulge area at 3 days after hair with or without topical H2O2 treatments (H2O2). H2O2 triggered the stress responsive (SR) type of ACDs. b, Representative IF images of KRT15, COL17A1 and ITGA6 in the bulge area of control, Col17a1 cKO and Itga6 cKO mice at 11 weeks old. A deficiency of Col17a1 or Itga6 affected the stability of the expression of each protein in the bulge area. c, Quantification of percent cells with polarized localization in the Bg and sBg areas of Control (n = 9 mice, n = 43 cells), Col17a1 cKO (n = 7 mice, n = 25 cells), Itga6 cKO (n = 3 mice, n = 23 cells) and Col17a1:Itga6 dcKO (n = 3 mice, n = 26 cells) mice. Col17a1 and/or Itga6 deficiency significantly abolished the polarized expression. d, IF images of KRT75 and KRT10 in control, Col17a1 cKO and Itga6 cKO mice at 3d. Col17a1 or Itga6 deficient HFSCs differentiated into KRT10+ cells, but not KRT75+ cells through the SR type of ACDs. e, Heat map showing the log fold change of HFSC signature genes between RU486-treated control (Cont) and Col17a1 cKO telogen stage HFSCs (tHFSCs) from microarray data. Col17a1 cKO tHFSCs decreased the expression of HFSC signature genes. f, Quantification of LacZ+ HFSCs contribution in the Bg, SG, JZ and EPI after nonHC, 3xHC and 5xHCs (Cont, nonHC, n = 3 mice, n = 37 HFs; Cont, 3xHC, n = 3 mice, n = 27 HFs; Cont, 5xHC, n = 46 HFs; Col17a1 cKO, nonHC, n = 3 mice, n = 33 HFs; Col17a1 cKO, 3xHC, n = 2 mice, n = 27 HFs; Col17a1 cKO, 5xHC, n = 3 mice, n = 68 HFs). LacZ-labeled control HFSCs retained the Bg, whereas Col17a1 and/or Itga6 deficient HFSCs dose-dependently retained SG, JZ and EPI. g, Representative TEM images of the basement membrane structure of control, Col17a1 and Itga6 cKO mice. Right panels show schematics for rete ridge-like structures in the basement membrane. h, Quantification of the depth of rete ridge-like structures (n = 3 mice, 27–34 images in each group). A deficiency of Col17a1 or Itga6 significantly decreased the depth of rete ridge-like structures. White dashed line, basement membrane; white line and fan-shaped line and upper number, spindle angle against basal cell layer (o); green(control)/red(IR or cKOs) dashed line, dividing cells. Error bars, means ± SEM; Chi-square test (c), one-way ANOVA with Tukey’s post-hoc test (f) or Dunnett’s post-hoc test (h).
Extended Data Fig. 3 Phenotypic consequence of an HFSC specific deficiency of the aPKCλ gene.
a, Representative IF images of KRT15 and aPKCλ in the bulge area and epidermis of control and aPKCλ cKO mice at 11 weeks of age. b, Quantification of the percent of aPKCλ deficient HFs in control and in aPKCλ cKO mice (control, n = 14 images; aPKCλ cKO, n = 14 images). aPKCλ cKO mice have a significantly decreased expression of aPKCλ in the bulge area. c, Quantification of percent cells with polarized localization in the Bg and sBg areas of Control (n = 9 mice, n = 43 cells), aPKCλ cKO (n = 4 mice, n = 25 cells) mice. aPKCλ deficiency significantly abolished the polarized localization. d, IF images of KRT15 and KRT1 in the dorsal skin of 11 wo control and aPKCλ cKO mice at 3 days after HC induction. e, Quantification of the percentage of KRT1+ cells per each HF after HC induction (control, n = 34 images; aPKCλ cKO, n = 8 images). aPKCλ cKO significantly increased the percentage of KRT1+ cells per each HF. f, IF images of KRT1 and KRT75 in control or in aPKCλ cKO mice at 3 days after HC induction. aPKCλ deficient-HFSCs differentiate into KRT1+ cells through SR type of ACDs. g, IF images of LacZ and LRIG1 in the dorsal skin of 11 wo control and aPKCλ cKO mice at 3 days after HC induction. LacZ+LRIG1+ (arrows) in HFSCs were found in aPKCλ cKO mice. h, Gene set enrichment analysis (GSEA) profile with ‘RAMALHO STEMNESS UP’ molecular signature in anagen stage (aHFSC) fraction between Cont and aPKCλ cKO mice. i, Heat map showing the log fold expression changes (logFC) of HFSC signature genes between RU486-treated control (Cont) and aPKCλ cKO telogen stage HFSCs (tHFSCs) from microarray data. aPKCλ cKO tHFSCs had decreased expression of HFSC signature genes excluding the Col17a1 gene. j, Heat map showing the log fold change of epidermal differentiation-related genes between Cont and aPKCλ cKO aHFSCs from microarray data. aPKCλ cKO aHFSCs had increased expression of epidermal differentiation-related genes including NOTCH related genes. k, IF images of NOTCH1, KRT1 and KRT15 in 11 wo Cont and aPKCλ cKO mice. NOTCH1+KRT1+ (arrows) in KRT15+ HFSCs were found in 11 wo aPKCλ cKO mice at 3 days after HC induction. l, IF images of c-MYC and KRT15 in 11 wo Cont and aPKCλ cKO mice. c-MYC+ cells (arrows) in KRT15+ HFSCs were found in 11 wo aPKCλ cKO mice at 3 days after HC induction. m, IH images of LacZ and Oil red O staining in 15 wo Cont and aPKCλ cKO mice. HFSC-derived LacZ+ cells (arrows) migrated into the epidermis together with SGs in 15 wo aPKCλ cKO mice at 1 month after HC induction. n, Schematic for experimental hypothesis of the interaction between the loss of aPKCλ and hemidesmosomes. o, p, q, Representative IF images of KRT15, COL17A1 (o), ITGA6 (p) and ITGB4 (q) in the bulge area of control and aPKCλ cKO mice at 11 weeks old. A deficiency of aPKCλ did not affect the expression level of HD components, COL17A1, ITGA6 and ITGB4. Error bars, means ± SEM; White dashed line, basement membrane; white line and fan-shaped line and upper number, spindle angle against basal cell layer (o); green(control)/red(cKOs) dashed line, dividing HFSCs. Chi-square test (c) or two-tailed Student’s t test (b, e).
Extended Data Fig. 4 Hemidesmosomal components stabilize cell polarity regulators.
a, Representative IF images of aPKCλ, COL17A1, CDC42, NUMB and PARD3 in HaCaT cells. Arrows indicates the co-localization between COL17A1 and cell polarity-related proteins. b, IF images of COL17A1 and PARD3 in si Cont (control) and in COL17A1 siRNA transduced NHEKs at 72 hr. c, IF images of PARD3, F-actin, NUMB and Plectin in si Cont (control) and in ITGA6 siRNA transduced NHEKs at 72 hr. KD of COL17A1 or ITGA6 decreased the polarized localization of cell polarity-related proteins in NHEKs (b, c). d, e, Quantitative RT-PCR analysis of mRNA expression levels of COL17A1 (d) and aPKCλ (e) in HaCaT cells at 96 hr after transfection of the indicated siRNA. f, g, Western blot hybridization analysis of COL17A1, aPKCλ, NUMB and TUBULIN in sh SCR or sh COL17A1 expressing stable HaCaT cell lines (f) or in HaCaT cells at 72 hr after transfection with si Cont or COL17A1 siRNA or pCMV-COL17A1 (g). h, j, IF images of COL17A1, aPKCλ, PARD3, NUMB and CDC42 in pCMV-COL17A1 transduced HaCaT cells (h) and NHEKs (j). i, k, IF images of ITGA6 and NUMB or aPKCλ in ITGA6-GFP over-expressing HaCaT cells (i) and NHEKs (k). OE of COL17A1 or ITGA6 provoked the increased expression of cell polarity-related proteins in the cortical region in HaCaT cells and in NHEKs (h, i, j, k). l, Immunoprecipitation analysis of the interaction between COL17A1 and aPKCλ in NHEK lysates. m, Immunoprecipitation analysis of the interaction between COL17A1 and aPKCλ in HaCaT cell lysates. Error bars, means ± SD; one-way ANOVA with Holm Sidak post-hoc test (d, e).
Extended Data Fig. 5 Establishment of an in vitro keratinocyte competition assay.
a, b, IF images of COL17A1 (a) or aPKCλ (b) in HaCaT cells at 72 hr after transfection of EmGFP-tagged sh SCRAMBLE (SCR) or sh COL17A1 or sh aPKCλ expressing vectors. COL17A1 or aPKCλ expression were decreased in EGFP tagged HaCaT cells (dashed lines), respectively. c, Western blot hybridization analysis of aPKCλ in EmGFP-tagged sh SCR or sh aPKCλ expressing stable HaCaT cell lines. d, Quantification of cell proliferation rates in EGFP tagged sh SCR or sh COL17A1 or sh aPKCλ stable HaCaT cell lines (n = 4 wells each lines). Cell proliferation was not significantly different in any of these cell lines. e, Western blot hybridization analysis of COL17A1 and EmGFP in the doxycycline (Dox) inducible EmGFP tagged sh SCR or sh COL17A1 expressing stable HaCaT cell lines with or without Dox. Dox treatments induce EmGFP and COL17A1 knockdown. f, g, h, IF images of COL17A1, EmGFP and KRT10 after 3D-coculture with sh SCR or sh COL17A1 stable HaCaT cell lines and wild-type HaCaT cells at ratios of 10:0 (f) and 1:10 (g, h) with DOX from -3 days (h) or 1 day (f, g) after plating. Arrows, EmGFP+ basal cells; White dashed line, basement membrane. i, Quantification of EmGFP+ cell contribution in the basal cell layer (sh SCR, Dox 1d, n = 15; sh COL17A1, Dox 1d, n = 15; sh SCR, Dox -3d, n = 10; sh COL17A1, Dox -3d, n = 11 images). COL17A1 KD cells were significantly eliminated by wild-type cells and did not depend on their ability to attach to the base of the dish. Error bars, means ± SEM; Kruskal-Wallis test with Dunn’s post hoc test (d); two tailed Student’s t-test (i).
Extended Data Fig. 6 Physiological aging and/or the DNA damage response triggers the destabilization of cell polarity-related proteins in HFSCs.
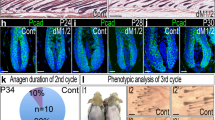
a, Heat map showing the log fold expression changes (logFC) of cell polarity-related genes between RU486-treated wild-type control (Cont) and aged (25mo) anagen stage HFSCs (aHFSCs) from microarray data. b, GSEA enrichment score curve of control (12 wo) versus aged (25 mo) HFSCs with the gene set for ‘CDC42RAC1 PATHWAY’ from microarray data. c, GSEA enrichment score curve of young (8 wo) versus aged (22–25 mo) HFSCs with the gene set for cell polarity-related genes from microarray data (GSE72683). d, Heat map showing the logFC of HFSC signature genes between young (8 wo) and aged (25 mo) anagen stage HFSCs (aHFSCs) from microarray data (GSE72683 from Matsumura et al. Science 2016). Aged aHFSCs had decreased expression of cell polarity-related genes. e, IF images of NUMB, CDC42 and PARD3 in the Bg area of young (8 wo) and of aged (24 mo) mice at 3 days after plucking (3d). f, Western blot hybridization analysis of COL17A1, CDC42, aPKCλ, NUMB, RAC1, PARD3 and TUBULIN in total epidermal lysates from young (8 wo) and from aged (24 mo) at 3d. Physiological aging destabilizes the cell polarity-related proteins. g,Venn diagram showing overlaps between 2> fold increased genes in cont vs aged HFSCs, 2> fold increased genes in cont vs Col17a1 cKO HFSCs, and 2> fold increased genes in cont vs aPKCλ cKO HFSCs. h, Venn diagram showing overlaps between 2> fold decreased genes in cont vs aged HFSCs, 2> fold decreased genes in cont vs Col17a1 cKO HFSCs, and 2> fold decreased genes in cont vs aPKCλ cKO HFSCs. i, IF images of ITGA6, COL17A1, NUMB and PARD3 in the bulge area of 12 mo control and of XPDTTD/TTD mutant mice. XPDTTD/TTD mutant HFSCs had decreased expression of cell polarity-related proteins. j, IF images of KRT15 and NUMB at 7 days after 10 Gy X-ray irradiation. hCOL17A1 transgenes rescued the IR-mediated instability of NUMB expression. k, IF images of KRT15 and aPKCλ in the Bg area from wild-type control or hCOL17A1 tg young (8 wo) and aged (25 mo) mice. hCOL17A1 transgenes rescued the age-associated instability of cell polarity-related proteins. l, TEM analysis of HD structure of young (8 wo), aged (25 mo) and aged (25 mo) hCOL17A1 transgenic mice. Right panels indicate magnified views of areas marked by red arrows on the left. Blue arrows indicate HDs. Small insets indicate magnified views of HDs (Red dotted lines). Bg, bulge area; HS, hair shaft. m, Quantification of HD number (8w, n = 3 HFs, n = 34 images; 25mo, n = 3 HFs, n = 16 images, 25mo tg, n = 2 HFs, n = 15 images). Expression of the hCOL17A1 transgene significantly rescued aged-associated immaturity and the reduced number of HDs. Error bars, means ± SEM; one-way ANOVA with Dunnett’s post-hoc test (m).
Extended Data Fig. 7 Schematic model of the distinct types of stem cell divisions determine HF homeostasis and aging.
Young HFSCs undergo symmetric cell divisions (SCDs) and asymmetric cell divisions (ACDs) to generate new bulge cells for their self-renewal and expansion. Whereas aged HFSCs provoke hemidesmosomal instability including COL17A1 and undergo stress response (SR) type ACDs to induce epidermal differentiation that triggers their delamination, thereby causing stepwise miniaturization of HFs and hair thinning and loss.
Supplementary information
Supplementary Information
Supplementary Table 1 and Fig. 1.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots and/or gels.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots and/or gels.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots and/or gels.
Rights and permissions
About this article
Cite this article
Matsumura, H., Liu, N., Nanba, D. et al. Distinct types of stem cell divisions determine organ regeneration and aging in hair follicles. Nat Aging 1, 190–204 (2021). https://doi.org/10.1038/s43587-021-00033-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43587-021-00033-7