Abstract
Bayesian statistics is an approach to data analysis based on Bayes’ theorem, where available knowledge about parameters in a statistical model is updated with the information in observed data. The background knowledge is expressed as a prior distribution and combined with observational data in the form of a likelihood function to determine the posterior distribution. The posterior can also be used for making predictions about future events. This Primer describes the stages involved in Bayesian analysis, from specifying the prior and data models to deriving inference, model checking and refinement. We discuss the importance of prior and posterior predictive checking, selecting a proper technique for sampling from a posterior distribution, variational inference and variable selection. Examples of successful applications of Bayesian analysis across various research fields are provided, including in social sciences, ecology, genetics, medicine and more. We propose strategies for reproducibility and reporting standards, outlining an updated WAMBS (when to Worry and how to Avoid the Misuse of Bayesian Statistics) checklist. Finally, we outline the impact of Bayesian analysis on artificial intelligence, a major goal in the next decade.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 1 digital issues and online access to articles
$99.00 per year
only $99.00 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
Change history
03 February 2021
A Correction to this paper has been published: https://doi.org/10.1038/s43586-021-00017-2.
References
Bayes, M. & Price, M. LII. An essay towards solving a problem in the doctrine of chances. By the late Rev. Mr. Bayes, F. R. S. communicated by Mr. Price, in a letter to John Canton, A. M. F. R. S. Philos. Trans. R Soc. Lond. B Biol. Sci. 53, 370–418 (1997).
Laplace, P. S. Essai Philosophique sur les Probabilities (Courcier, 1814).
König, C. & van de Schoot, R. Bayesian statistics in educational research: a look at the current state of affairs. Educ. Rev. https://doi.org/10.1080/00131911.2017.1350636 (2017).
van de Schoot, R., Winter, S., Zondervan-Zwijnenburg, M., Ryan, O. & Depaoli, S. A systematic review of Bayesian applications in psychology: the last 25 years. Psychol. Methods 22, 217–239 (2017).
Ashby, D. Bayesian statistics in medicine: a 25 year review. Stat. Med. 25, 3589–3631 (2006).
Rietbergen, C., Debray, T. P. A., Klugkist, I., Janssen, K. J. M. & Moons, K. G. M. Reporting of Bayesian analysis in epidemiologic research should become more transparent. J. Clin. Epidemiol. https://doi.org/10.1016/j.jclinepi.2017.04.008 (2017).
Spiegelhalter, D. J., Myles, J. P., Jones, D. R. & Abrams, K. R. Bayesian methods in health technology assessment: a review. Health Technol. Assess. https://doi.org/10.3310/hta4380 (2000).
Kruschke, J. K., Aguinis, H. & Joo, H. The time has come: Bayesian methods for data analysis in the organizational sciences. Organ. Res. Methods 15, 722–752 (2012).
Smid, S. C., McNeish, D., Miočević, M. & van de Schoot, R. Bayesian versus frequentist estimation for structural equation models in small sample contexts: a systematic review. Struct. Equ. Modeling 27, 131–161 (2019).
Rupp, A. A., Dey, D. K. & Zumbo, B. D. To Bayes or not to Bayes, from whether to when: applications of Bayesian methodology to modeling. Struct. Equ. Modeling 11, 424–451 (2004).
van de Schoot, R., Yerkes, M. A., Mouw, J. M. & Sonneveld, H. What took them so long? Explaining PhD delays among doctoral candidates. PloS ONE 8, e68839 (2013).
van de Schoot, R. Online stats training. Zenodo https://zenodo.org/communities/stats_training (2020).
Heo, I. & van de Schoot, R. Tutorial: advanced Bayesian regression in JASP. Zenodo https://doi.org/10.5281/zenodo.3991325 (2020).
O’Hagan, A. et al. Uncertain Judgements: Eliciting Experts’ Probabilities (Wiley, 2006). This book presents a great collection of information with respect to prior elicitation, and includes elicitation techniques, summarizes potential pitfalls and describes examples across a wide variety of disciplines.
Howard, G. S., Maxwell, S. E. & Fleming, K. J. The proof of the pudding: an illustration of the relative strengths of null hypothesis, meta-analysis, and Bayesian analysis. Psychol. Methods 5, 315–332 (2000).
Veen, D., Stoel, D., Zondervan-Zwijnenburg, M. & van de Schoot, R. Proposal for a five-step method to elicit expert judgement. Front. Psychol. 8, 2110 (2017).
Johnson, S. R., Tomlinson, G. A., Hawker, G. A., Granton, J. T. & Feldman, B. M. Methods to elicit beliefs for Bayesian priors: a systematic review. J. Clin. Epidemiol. 63, 355–369 (2010).
Morris, D. E., Oakley, J. E. & Crowe, J. A. A web-based tool for eliciting probability distributions from experts. Environ. Model. Softw. https://doi.org/10.1016/j.envsoft.2013.10.010 (2014).
Garthwaite, P. H., Al-Awadhi, S. A., Elfadaly, F. G. & Jenkinson, D. J. Prior distribution elicitation for generalized linear and piecewise-linear models. J. Appl. Stat. 40, 59–75 (2013).
Elfadaly, F. G. & Garthwaite, P. H. Eliciting Dirichlet and Gaussian copula prior distributions for multinomial models. Stat. Comput. 27, 449–467 (2017).
Veen, D., Egberts, M. R., van Loey, N. E. E. & van de Schoot, R. Expert elicitation for latent growth curve models: the case of posttraumatic stress symptoms development in children with burn injuries. Front. Psychol. 11, 1197 (2020).
Runge, A. K., Scherbaum, F., Curtis, A. & Riggelsen, C. An interactive tool for the elicitation of subjective probabilities in probabilistic seismic-hazard analysis. Bull. Seismol. Soc. Am. 103, 2862–2874 (2013).
Zondervan-Zwijnenburg, M., van de Schoot-Hubeek, W., Lek, K., Hoijtink, H. & van de Schoot, R. Application and evaluation of an expert judgment elicitation procedure for correlations. Front. Psychol. https://doi.org/10.3389/fpsyg.2017.00090 (2017).
Cooke, R. M. & Goossens, L. H. J. TU Delft expert judgment data base. Reliab. Eng. Syst. Saf. 93, 657–674 (2008).
Hanea, A. M., Nane, G. F., Bedford, T. & French, S. Expert Judgment in Risk and Decision Analysis (Springer, 2020).
Dias, L. C., Morton, A. & Quigley, J. Elicitation (Springer, 2018).
Ibrahim, J. G., Chen, M. H., Gwon, Y. & Chen, F. The power prior: theory and applications. Stat. Med. 34, 3724–3749 (2015).
Rietbergen, C., Klugkist, I., Janssen, K. J., Moons, K. G. & Hoijtink, H. J. Incorporation of historical data in the analysis of randomized therapeutic trials. Contemp. Clin. Trials 32, 848–855 (2011).
van de Schoot, R. et al. Bayesian PTSD-trajectory analysis with informed priors based on a systematic literature search and expert elicitation. Multivariate Behav. Res. 53, 267–291 (2018).
Berger, J. The case for objective Bayesian analysis. Bayesian Anal. 1, 385–402 (2006). This discussion of objective Bayesian analysis includes criticisms of the approach and a personal perspective on the debate on the value of objective Bayesian versus subjective Bayesian analysis.
Brown, L. D. In-season prediction of batting averages: a field test of empirical Bayes and Bayes methodologies. Ann. Appl. Stat. https://doi.org/10.1214/07-AOAS138 (2008).
Candel, M. J. & Winkens, B. Performance of empirical Bayes estimators of level-2 random parameters in multilevel analysis: a Monte Carlo study for longitudinal designs. J. Educ. Behav. Stat. 28, 169–194 (2003).
van der Linden, W. J. Using response times for item selection in adaptive testing. J. Educ. Behav. Stat. 33, 5–20 (2008).
Darnieder, W. F. Bayesian Methods for Data-Dependent Priors (The Ohio State Univ., 2011).
Richardson, S. & Green, P. J. On Bayesian analysis of mixtures with an unknown number of components (with discussion). J. R. Stat. Soc. Series B 59, 731–792 (1997).
Wasserman, L. Asymptotic inference for mixture models by using data-dependent priors. J. R. Stat. Soc. Series B 62, 159–180 (2000).
Muthen, B. & Asparouhov, T. Bayesian structural equation modeling: a more flexible representation of substantive theory. Psychol. Methods 17, 313–335 (2012).
van de Schoot, R. et al. Facing off with Scylla and Charybdis: a comparison of scalar, partial, and the novel possibility of approximate measurement invariance. Front. Psychol. 4, 770 (2013).
Smeets, L. & van de Schoot, R. Code for the ShinyApp to determine the plausible parameter space for the PhD-delay data (version v1.0). Zenodo https://doi.org/10.5281/zenodo.3999424 (2020).
Chung, Y., Gelman, A., Rabe-Hesketh, S., Liu, J. & Dorie, V. Weakly informative prior for point estimation of covariance matrices in hierarchical models. J. Educ. Behav. Stat. 40, 136–157 (2015).
Gelman, A., Jakulin, A., Pittau, M. G. & Su, Y.-S. A weakly informative default prior distribution for logistic and other regression models. Ann. Appl. Stat. 2, 1360–1383 (2008).
Gelman, A., Carlin, J. B., Stern, H. S. & Rubin, D. B. Bayesian Data Analysis Vol. 2 (Chapman&HallCRC, 2004).
Jeffreys, H. Theory of Probability Vol. 3 (Clarendon, 1961).
Seaman III, J. W., Seaman Jr, J. W. & Stamey, J. D. Hidden dangers of specifying noninformative priors. Am. Stat. 66, 77–84 (2012).
Gelman, A. Prior distributions for variance parameters in hierarchical models (comment on article by Browne and Draper). Bayesian Anal. 1, 515–534 (2006).
Lambert, P. C., Sutton, A. J., Burton, P. R., Abrams, K. R. & Jones, D. R. How vague is vague? A simulation study of the impact of the use of vague prior distributions in MCMC using WinBUGS. Stat. Med. 24, 2401–2428 (2005).
Depaoli, S. Mixture class recovery in GMM under varying degrees of class separation: frequentist versus Bayesian estimation. Psychol. Methods 18, 186–219 (2013).
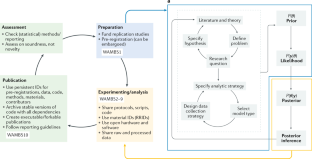
Depaoli, S. & van de Schoot, R. Improving transparency and replication in Bayesian statistics: the WAMBS-Checklist. Psychol. Methods 22, 240 (2017). This article describes, in a step-by-step manner, the various points that need to be checked when estimating a model using Bayesian statistics. It can be used as a guide for implementing Bayesian methods.
van Erp, S., Mulder, J. & Oberski, D. L. Prior sensitivity analysis in default Bayesian structural equation modeling. Psychol. Methods 23, 363–388 (2018).
McNeish, D. On using Bayesian methods to address small sample problems. Struct. Equ. Modeling 23, 750–773 (2016).
van de Schoot, R. & Miocević, M. Small Sample Size Solutions: A Guide for Applied Researchers and Practitioners (Taylor & Francis, 2020).
Schuurman, N. K., Grasman, R. P. & Hamaker, E. L. A comparison of inverse-Wishart prior specifications for covariance matrices in multilevel autoregressive models. Multivariate Behav. Res. 51, 185–206 (2016).
Liu, H., Zhang, Z. & Grimm, K. J. Comparison of inverse Wishart and separation-strategy priors for Bayesian estimation of covariance parameter matrix in growth curve analysis. Struct. Equ. Modeling 23, 354–367 (2016).
Ranganath, R. & Blei, D. M. Population predictive checks. Preprint at https://arxiv.org/abs/1908.00882 (2019).
Daimon, T. Predictive checking for Bayesian interim analyses in clinical trials. Contemp. Clin. Trials 29, 740–750 (2008).
Box, G. E. Sampling and Bayes’ inference in scientific modelling and robustness. J. R. Stat. Soc. Ser. A 143, 383–404 (1980).
Gabry, J., Simpson, D., Vehtari, A., Betancourt, M. & Gelman, A. Visualization in Bayesian workflow. J. R. Stat. Soc. Ser. A 182, 389–402 (2019).
Silverman, B. W. Density Estimation for Statistics and Data Analysis Vol. 26 (CRC, 1986).
Nott, D. J., Drovandi, C. C., Mengersen, K. & Evans, M. Approximation of Bayesian predictive p-values with regression ABC. Bayesian Anal. 13, 59–83 (2018).
Evans, M. & Moshonov, H. in Bayesian Statistics and its Applications 145–159 (Univ. of Toronto, 2007).
Evans, M. & Moshonov, H. Checking for prior–data conflict. Bayesian Anal. 1, 893–914 (2006).
Evans, M. & Jang, G. H. A limit result for the prior predictive applied to checking for prior–data conflict. Stat. Probab. Lett. 81, 1034–1038 (2011).
Young, K. & Pettit, L. Measuring discordancy between prior and data. J. R. Stat. Soc. Series B Methodol. 58, 679–689 (1996).
Kass, R. E. & Raftery, A. E. Bayes factors. J. Am. Stat. Assoc. 90, 773–795 (1995). This article provides an extensive discussion of Bayes factors with several examples.
Bousquet, N. Diagnostics of prior–data agreement in applied Bayesian analysis. J. Appl. Stat. 35, 1011–1029 (2008).
Veen, D., Stoel, D., Schalken, N., Mulder, K. & van de Schoot, R. Using the data agreement criterion to rank experts’ beliefs. Entropy 20, 592 (2018).
Nott, D. J., Xueou, W., Evans, M. & Englert, B. Checking for prior–data conflict using prior to posterior divergences. Preprint at https://arxiv.org/abs/1611.00113 (2016).
Lek, K. & van de Schoot, R. How the choice of distance measure influences the detection of prior–data conflict. Entropy 21, 446 (2019).
O’Hagan, A. Bayesian statistics: principles and benefits. Frontis 3, 31–45 (2004).
Etz, A. Introduction to the concept of likelihood and its applications. Adv. Methods Practices Psychol. Sci. 1, 60–69 (2018).
Pawitan, Y. In All Likelihood: Statistical Modelling and Inference Using Likelihood (Oxford Univ. Press, 2001).
Gelman, A., Simpson, D. & Betancourt, M. The prior can often only be understood in the context of the likelihood. Entropy 19, 555 (2017).
Aczel, B. et al. Discussion points for Bayesian inference. Nat. Hum. Behav. 4, 561–563 (2020).
Gelman, A. et al. Bayesian Data Analysis (CRC, 2013).
Greco, L., Racugno, W. & Ventura, L. Robust likelihood functions in Bayesian inference. J. Stat. Plan. Inference 138, 1258–1270 (2008).
Shyamalkumar, N. D. in Robust Bayesian Analysis Lecture Notes in Statistics Ch. 7, 127–143 (Springer, 2000).
Agostinelli, C. & Greco, L. A weighted strategy to handle likelihood uncertainty in Bayesian inference. Comput. Stat. 28, 319–339 (2013).
Rubin, D. B. Bayesianly justifiable and relevant frequency calculations for the applied statistician. Ann. Stat. 12, 1151–1172 (1984).
Gelfand, A. E. & Smith, A. F. M. Sampling-based approaches to calculating marginal densities. J. Am. Stat. Assoc. 85, 398–409 (1990). This seminal article identifies MCMC as a practical approach for Bayesian inference.
Geyer, C. J. Markov chain Monte Carlo maximum likelihood. IFNA http://hdl.handle.net/11299/58440 (1991).
van de Schoot, R., Veen, D., Smeets, L., Winter, S. D. & Depaoli, S. in Small Sample Size Solutions: A Guide for Applied Researchers and Practitioners Ch. 3 (eds van de Schoot, R. & Miocevic, M.) 30–49 (Routledge, 2020).
Veen, D. & Egberts, M. in Small Sample Size Solutions: A Guide for Applied Researchers and Practitioners Ch. 4 (eds van de Schoot, R. & Miocevic, M.) 50–70 (Routledge, 2020).
Robert, C. & Casella, G. Monte Carlo Statistical Methods (Springer Science & Business Media, 2013).
Geman, S. & Geman, D. Stochastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE Trans. Pattern Anal. Mach. Intell. 6, 721–741 (1984).
Metropolis, N., Rosenbluth, A. W., Rosenbluth, M. N., Teller, A. H. & Teller, E. Equation of state calculations by fast computing machines. J. Chem. Phys. 21, 1087–1092 (1953).
Hastings, W. K. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57, 97–109 (1970).
Duane, S., Kennedy, A. D., Pendleton, B. J. & Roweth, D. Hybrid Monte Carlo. Phys. Lett. B 195, 216–222 (1987).
Tanner, M. A. & Wong, W. H. The calculation of posterior distributions by data augmentation. J. Am. Stat. Assoc. 82, 528–540 (1987). This article explains how to use data augmentation when direct computation of the posterior density of the parameters of interest is not possible.
Gamerman, D. & Lopes, H. F. Markov Chain Monte Carlo: Stochastic Simulation for Bayesian Inference (CRC, 2006).
Brooks, S. P., Gelman, A., Jones, G. & Meng, X.-L. Handbook of Markov Chain Monte Carlo (CRC, 2011). This book presents a comprehensive review of MCMC and its use in many different applications.
Gelman, A. Burn-in for MCMC, why we prefer the term warm-up. Satistical Modeling, Causal Inference, and Social Science https://statmodeling.stat.columbia.edu/2017/12/15/burn-vs-warm-iterative-simulation-algorithms/ (2017).
Gelman, A. & Rubin, D. B. Inference from iterative simulation using multiple sequences. Stat. Sci. 7, 457–511 (1992).
Brooks, S. P. & Gelman, A. General methods for monitoring convergence of iterative simulations. J. Comput. Graph. Stat. 7, 434–455 (1998).
Roberts, G. O. Markov chain concepts related to sampling algorithms. Markov Chain Monte Carlo in Practice 57, 45–58 (1996).
Vehtari, A., Gelman, A., Simpson, D., Carpenter, B. & Bürkner, P. Rank-normalization, folding, and localization: an improved \(\hat{R}\) for assessing convergence of MCMC. Preprint at https://arxiv.org/abs/1903.08008 (2020).
Bürkner, P.-C. Advanced Bayesian multilevel modeling with the R package brms. Preprint at https://arxiv.org/abs/1705.11123 (2017).
Merkle, E. C. & Rosseel, Y. blavaan: Bayesian structural equation models via parameter expansion. Preprint at https://arxiv.org/abs/1511.05604 (2015).
Carpenter, B. et al. Stan: a probabilistic programming language. J. Stat. Softw. https://doi.org/10.18637/jss.v076.i01 (2017).
Blei, D. M., Kucukelbir, A. & McAuliffe, J. D. Variational inference: a review for statisticians. J. Am. Stat. Assoc. 112, 859–877 (2017). This recent review of variational inference methods includes stochastic variants that underpin popular approximate Bayesian inference methods for large data or complex modelling problems.
Minka, T. P. Expectation propagation for approximate Bayesian inference. Preprint at https://arxiv.org/abs/1301.2294 (2013).
Hoffman, M. D., Blei, D. M., Wang, C. & Paisley, J. Stochastic variational inference. J. Mach. Learn. Res. 14, 1303–1347 (2013).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. Preprint at https://arxiv.org/abs/1412.6980 (2014).
Li, Y., Hernández-Lobato, J. M. & Turner, R. E. Stochastic expectation propagation. Adv. Neural Inf. Process. Syst. 28, 2323–2331 (2015).
Liang, F., Paulo, R., Molina, G., Clyde, M. A. & Berger, J. O. Mixtures of g priors for Bayesian variable selection. J. Am. Stat. Assoc. 103, 410–423 (2008).
Forte, A., Garcia-Donato, G. & Steel, M. Methods and tools for Bayesian variable selection and model averaging in normal linear regression. Int. Stat.Rev. 86, 237–258 (2018).
Mitchell, T. J. & Beauchamp, J. J. Bayesian variable selection in linear regression. J. Am. Stat. Assoc. 83, 1023–1032 (1988).
George, E. J. & McCulloch, R. E. Variable selection via Gibbs sampling. J. Am. Stat. Assoc. 88, 881–889 (1993). This article popularizes the use of spike-and-slab priors for Bayesian variable selection and introduces MCMC techniques to explore the model space.
Ishwaran, H. & Rao, J. S. Spike and slab variable selection: frequentist and Bayesian strategies. Ann. Stat. 33, 730–773 (2005).
Bottolo, L. & Richardson, S. Evolutionary stochastic search for Bayesian model exploration. Bayesian Anal. 5, 583–618 (2010).
Ročková, V. & George, E. I. EMVS: the EM approach to Bayesian variable selection. J. Am. Stat. Assoc. 109, 828–846 (2014).
Park, T. & Casella, G. The Bayesian lasso. J. Am. Stat. Assoc. 103, 681–686 (2008).
Carvalho, C. M., Polson, N. G. & Scott, J. G. The horseshoe estimator for sparse signals. Biometrika 97, 465–480 (2010).
Polson, N. G. & Scott, J. G. Shrink globally, act locally: sparse Bayesian regularization and prediction. Bayesian Stat. 9, 105 (2010). This article provides a unified framework for continuous shrinkage priors, which allow global sparsity while controlling the amount of regularization for each regression coefficient.
Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Series B 58, 267–288 (1996).
Van Erp, S., Oberski, D. L. & Mulder, J. Shrinkage priors for Bayesian penalized regression. J. Math. Psychol. 89, 31–50 (2019).
Brown, P. J., Vannucci, M. & Fearn, T. Multivariate Bayesian variable selection and prediction. J. R. Stat. Soc. Series B 60, 627–641 (1998).
Lee, K. H., Tadesse, M. G., Baccarelli, A. A., Schwartz, J. & Coull, B. A. Multivariate Bayesian variable selection exploiting dependence structure among outcomes: application to air pollution effects on DNA methylation. Biometrics 73, 232–241 (2017).
Frühwirth-Schnatter, S. & Wagner, H. Stochastic model specification search for Gaussian and partially non-Gaussian state space models. J. Econom. 154, 85–100 (2010).
Scheipl, F., Fahrmeir, L. & Kneib, T. Spike-and-slab priors for function selection in structured additive regression models. J. Am. Stat. Assoc. 107, 1518–1532 (2012).
Tadesse, M. G., Sha, N. & Vannucci, M. Bayesian variable selection in clustering high dimensional data. J. Am. Stat. Assoc. https://doi.org/10.1198/016214504000001565 (2005).
Wang, H. Scaling it up: stochastic search structure learning in graphical models. Bayesian Anal. 10, 351–377 (2015).
Peterson, C. B., Stingo, F. C. & Vannucci, M. Bayesian inference of multiple Gaussian graphical models. J. Am. Stat. Assoc. 110, 159–174 (2015).
Li, F. & Zhang, N. R. Bayesian variable selection in structured high-dimensional covariate spaces with applications in genomics. J. Am. Stat. Assoc. 105, 1978–2002 (2010).
Stingo, F., Chen, Y., Tadesse, M. G. & Vannucci, M. Incorporating biological information into linear models: a Bayesian approach to the selection of pathways and genes. Ann. Appl. Stat. 5, 1202–1214 (2011).
Guan, Y. & Stephens, M. Bayesian variable selection regression for genome-wide association studies and other large-scale problems. Ann. Appl. Stat. 5, 1780–1815 (2011).
Bottolo, L. et al. GUESS-ing polygenic associations with multiple phenotypes using a GPU-based evolutionary stochastic search algorithm. PLoS Genetics 9, e1003657–e1003657 (2013).
Banerjee, S., Carlin, B. P. & Gelfand, A. E. Hierarchical Modeling and Analysis for Spatial Data (CRC, 2014).
Vock, L. F. B., Reich, B. J., Fuentes, M. & Dominici, F. Spatial variable selection methods for investigating acute health effects of fine particulate matter components. Biometrics 71, 167–177 (2015).
Penny, W. D., Trujillo-Barreto, N. J. & Friston, K. J. Bayesian fMRI time series analysis with spatial priors. Neuroimage 24, 350–362 (2005).
Smith, M., Pütz, B., Auer, D. & Fahrmeir, L. Assessing brain activity through spatial Bayesian variable selection. Neuroimage 20, 802–815 (2003).
Zhang, L., Guindani, M., Versace, F. & Vannucci, M. A spatio-temporal nonparametric Bayesian variable selection model of fMRI data for clustering correlated time courses. Neuroimage 95, 162–175 (2014).
Gorrostieta, C., Fiecas, M., Ombao, H., Burke, E. & Cramer, S. Hierarchical vector auto-regressive models and their applications to multi-subject effective connectivity. Front. Computat. Neurosci. 7, 159–159 (2013).
Chiang, S. et al. Bayesian vector autoregressive model for multi-subject effective connectivity inference using multi-modal neuroimaging data. Human Brain Mapping 38, 1311–1332 (2017).
Schad, D. J., Betancourt, M. & Vasishth, S. Toward a principled Bayesian workflow in cognitive science. Preprint at https://arxiv.org/abs/1904.12765 (2019).
Gelman, A., Meng, X.-L. & Stern, H. Posterior predictive assessment of model fitness via realized discrepancies. Stat. Sinica 6, 733–760 (1996).
Meng, X.-L. Posterior predictive p-values. Ann. Stat. 22, 1142–1160 (1994).
Asparouhov, T., Hamaker, E. L. & Muthén, B. Dynamic structural equation models. Struct. Equ. Modeling 25, 359–388 (2018).
Zhang, Z., Hamaker, E. L. & Nesselroade, J. R. Comparisons of four methods for estimating a dynamic factor model. Struct. Equ. Modeling 15, 377–402 (2008).
Hamaker, E., Ceulemans, E., Grasman, R. & Tuerlinckx, F. Modeling affect dynamics: state of the art and future challenges. Emot. Rev. 7, 316–322 (2015).
Meissner, P. wikipediatrend: Public Subject Attention via Wikipedia Page View Statistics. R package version 2.1.6. Peter Meissner https://CRAN.R-project.org/package=wikipediatrend (2020).
Veen, D. & van de Schoot, R. Bayesian analysis for PhD-delay dataset. OSF https://doi.org/10.17605/OSF.IO/JA859 (2020).
Harvey, A. C. & Peters, S. Estimation procedures for structural time series models. J. Forecast. 9, 89–108 (1990).
Taylor, S. J. & Letham, B. Forecasting at scale. Am. Stat. 72, 37–45 (2018).
Gopnik, A. & Bonawitz, E. Bayesian models of child development. Wiley Interdiscip. Rev. Cogn. Sci. 6, 75–86 (2015).
Gigerenzer, G. & Hoffrage, U. How to improve Bayesian reasoning without instruction: frequency formats. Psychol. Rev. 102, 684 (1995).
Slovic, P. & Lichtenstein, S. Comparison of Bayesian and regression approaches to the study of information processing in judgment. Organ. Behav. Hum. Perform. 6, 649–744 (1971).
Bolt, D. M., Piper, M. E., Theobald, W. E. & Baker, T. B. Why two smoking cessation agents work better than one: role of craving suppression. J. Consult. Clin. Psychol. 80, 54–65 (2012).
Billari, F. C., Graziani, R. & Melilli, E. Stochastic population forecasting based on combinations of expert evaluations within the Bayesian paradigm. Demography 51, 1933–1954 (2014).
Fallesen, P. & Breen, R. Temporary life changes and the timing of divorce. Demography 53, 1377–1398 (2016).
Hansford, T. G., Depaoli, S. & Canelo, K. S. Locating U.S. Solicitors General in the Supreme Court’s policy space. Pres. Stud. Q. 49, 855–869 (2019).
Phipps, D. J., Hagger, M. S. & Hamilton, K. Predicting limiting ‘free sugar’ consumption using an integrated model of health behavior. Appetite 150, 104668 (2020).
Depaoli, S., Rus, H. M., Clifton, J. P., van de Schoot, R. & Tiemensma, J. An introduction to Bayesian statistics in health psychology. Health Psychol. Rev. 11, 248–264 (2017).
Kruschke, J. K. Bayesian estimation supersedes the t test. J. Exp. Psychol. Gen. 142, 573–603 (2013).
Lee, M. D. How cognitive modeling can benefit from hierarchical Bayesian models. J. Math. Psychol. 55, 1–7 (2011).
Royle, J. & Dorazio, R. Hierarchical Modeling and Inference in Ecology (Academic, 2008).
Gimenez, O. et al. in Modeling Demographic Processes in Marked Populations Vol. 3 (eds Thomson D. L., Cooch E. G. & Conroy M. J.) 883–915 (Springer, 2009).
King, R., Morgan, B., Gimenez, O. & Brooks, S. P. Bayesian Analysis for Population Ecology (CRC, 2009).
Kéry, M. & Schaub, M. Bayesian Population Analysis using WinBUGS: A Hierarchical Perspective (Academic, 2011).
McCarthy, M. Bayesian Methods of Ecology 5th edn (Cambridge Univ. Press, 2012).
Korner-Nievergelt, F. et al. Bayesian Data Analysis in Ecology Using Linear Models with R, BUGS, and Stan (Academic, 2015).
Monnahan, C. C., Thorson, J. T. & Branch, T. A. Faster estimation of Bayesian models in ecology using Hamiltonian Monte Carlo. Methods Ecol. Evol. 8, 339–348 (2017).
Ellison, A. M. Bayesian inference in ecology. Ecol. Lett. 7, 509–520 (2004).
Choy, S. L., O’Leary, R. & Mengersen, K. Elicitation by design in ecology: using expert opinion to inform priors for Bayesian statistical models. Ecology 90, 265–277 (2009).
Kuhnert, P. M., Martin, T. G. & Griffiths, S. P. A guide to eliciting and using expert knowledge in Bayesian ecological models. Ecol. Lett. 13, 900–914 (2010).
King, R., Brooks, S. P., Mazzetta, C., Freeman, S. N. & Morgan, B. J. Identifying and diagnosing population declines: a Bayesian assessment of lapwings in the UK. J. R. Stat. Soc. Series C 57, 609–632 (2008).
Newman, K. et al. Modelling Population Dynamics (Springer, 2014).
Bachl, F. E., Lindgren, F., Borchers, D. L. & Illian, J. B. inlabru: an R package for Bayesian spatial modelling from ecological survey data. Methods Ecol. Evol. 10, 760–766 (2019).
King, R. & Brooks, S. P. On the Bayesian estimation of a closed population size in the presence of heterogeneity and model uncertainty. Biometrics 64, 816–824 (2008).
Saunders, S. P., Cuthbert, F. J. & Zipkin, E. F. Evaluating population viability and efficacy of conservation management using integrated population models. J. Appl. Ecol. 55, 1380–1392 (2018).
McClintock, B. T. et al. A general discrete-time modeling framework for animal movement using multistate random walks. Ecol. Monog. 82, 335–349 (2012).
Dennis, B., Ponciano, J. M., Lele, S. R., Taper, M. L. & Staples, D. F. Estimating density dependence, process noise, and observation error. Ecol. Monog. 76, 323–341 (2006).
Aeberhard, W. H., Mills Flemming, J. & Nielsen, A. Review of state-space models for fisheries science. Ann. Rev. Stat. Appl. 5, 215–235 (2018).
Isaac, N. J. B. et al. Data integration for large-scale models of species distributions. Trends Ecol Evol 35, 56–67 (2020).
McClintock, B. T. et al. Uncovering ecological state dynamics with hidden Markov models. Preprint at https://arxiv.org/abs/2002.10497 (2020).
King, R. Statistical ecology. Ann. Rev. Stat. Appl. 1, 401–426 (2014).
Fearnhead, P. in Handbook of Markov Chain Monte Carlo Ch. 21 (eds Brooks, S., Gelman, A., Jones, G.L. & Meng, X.L.) 513–529 (Chapman & Hall/CRC, 2011).
Andrieu, C., Doucet, A. & Holenstein, R. Particle Markov chain Monte Carlo methods. J. R. Stat. Soc. Series B 72, 269–342 (2010).
Knape, J. & de Valpine, P. Fitting complex population models by combining particle filters with Markov chain Monte Carlo. Ecology 93, 256–263 (2012).
Finke, A., King, R., Beskos, A. & Dellaportas, P. Efficient sequential Monte Carlo algorithms for integrated population models. J. Agric. Biol. Environ. Stat. 24, 204–224 (2019).
Stephens, M. & Balding, D. J. Bayesian statistical methods for genetic association studies. Nat. Rev. Genet 10, 681–690 (2009).
Mimno, D., Blei, D. M. & Engelhardt, B. E. Posterior predictive checks to quantify lack-of-fit in admixture models of latent population structure. Proc. Natl Acad. Sci. USA 112, E3441–3450 (2015).
Schaid, D. J., Chen, W. & Larson, N. B. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet. 19, 491–504 (2018).
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010).
Allen, N. E., Sudlow, C., Peakman, T., Collins, R. & Biobank, U. K. UK Biobank data: come and get it. Sci. Transl. Med. 6, 224ed224 (2014).
Cortes, A. et al. Bayesian analysis of genetic association across tree-structured routine healthcare data in the UK Biobank. Nat. Genet. 49, 1311–1318 (2017).
Argelaguet, R. et al. Multi-omics factor analysis — a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 14, e8124 (2018).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Yau, C. & Campbell, K. Bayesian statistical learning for big data biology. Biophys. Rev. 11, 95–102 (2019).
Vallejos, C. A., Marioni, J. C. & Richardson, S. BASiCS: Bayesian analysis of single-cell sequencing data. PLoS Comput. Biol. 11, e1004333 (2015).
Wang, J. et al. Data denoising with transfer learning in single-cell transcriptomics. Nat. Methods 16, 875–878 (2019).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
National Cancer Institute. The Cancer Genome Atlas. Qeios https://doi.org/10.32388/e1plqh (2020).
Kuipers, J. et al. Mutational interactions define novel cancer subgroups. Nat. Commun. 9, 4353 (2018).
Schwartz, R. & Schaffer, A. A. The evolution of tumour phylogenetics: principles and practice. Nat. Rev. Genet. 18, 213–229 (2017).
Munafò, M. R. et al. A manifesto for reproducible science. Nat. Hum. Behav. https://doi.org/10.1038/s41562-016-0021 (2017).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Lamprecht, A.-L. et al. Towards FAIR principles for research software. Data Sci. 3, 37–59 (2020).
Smith, A. M., Katz, D. S. & Niemeyer, K. E. Software citation principles. PeerJ Comput. Sci. 2, e86 (2016).
Clyburne-Sherin, A., Fei, X. & Green, S. A. Computational reproducibility via containers in psychology. Meta Psychol. https://doi.org/10.15626/MP.2018.892 (2019).
Lowenberg, D. Dryad & Zenodo: our path ahead. WordPress https://blog.datadryad.org/2020/03/10/dryad-zenodo-our-path-ahead/ (2020).
Nosek, B. A. et al. Promoting an open research culture. Science 348, 1422–1425 (2015).
Vehtari, A. & Ojanen, J. A survey of Bayesian predictive methods for model assessment, selection and comparison. Stat. Surv. 6, 142–228 (2012).
Abadi, M. et al. in USENIX Symposium on Operating Systems Design and Implementation (OSDI'16) 265–283 (USENIX Association, 2016).
Paszke, A. et al. in Advances in Neural Information Processing Systems (eds Wallach, H. et al.) 8026–8037 (Urran Associates, 2019).
Kingma, D. P. & Welling, M. An introduction to variational autoencoders. Preprint at https://arxiv.org/abs/1906.02691 (2019). This recent review of variational autoencoders encompasses deep generative models, the re-parameterization trick and current inference methods.
Higgins, I. et al. beta-VAE: learning basic visual concepts with a constrained variational framework. ICLR 2017 https://openreview.net/forum?id=Sy2fzU9gl (2017).
Märtens, K. & Yau, C. BasisVAE:tTranslation-invariant feature-level clustering with variational autoencoders. Preprint at https://arxiv.org/abs/2003.03462 (2020).
Liu, Q., Allamanis, M., Brockschmidt, M. & Gaunt, A. in Advances in Neural Information Processing Systems 31 (eds Bengio, S. et al.) 7795–7804 (Curran Associates, 2018).
Louizos, C., Shi, X., Schutte, K. & Welling, M. in Advances in Neural Information Processing Systems 8743-8754 (MIT Press, 2019).
Garnelo, M. et al. in Proceedings of the 35th International Conference on Machine Learning Vol. 80 (eds Dy, J. & Krause, A.) 1704–1713 (PMLR, 2018).
Kim, H. et al. Attentive neural processes. Preprint at https://arxiv.org/abs/1901.05761 (2019).
Rezende, D. & Mohamed, S. in Proceedings of the 32nd International Conference on Machine Learning Vol. 37 (eds Bach, F. & Blei, D.) 1530–1538 (PMLR, 2015).
Papamakarios, G., Nalisnick, E., Rezende, D. J., Mohamed, S. & Lakshminarayanan, B. Normalizing flows for probabilistic modeling and inference. Preprint at https://arxiv.org/abs/1912.02762 (2019).
Korshunova, I. et al. in Advances in Neural Information Processing Systems 31 (eds Bengio, S.et al.) 7190–7198 (Curran Associates, 2018).
Zhang, R., Li, C., Zhang, J., Chen, C. & Wilson, A. G. Cyclical stochastic gradient MCMC for Bayesian deep learning. Preprint at https://arxiv.org/abs/1902.03932 (2019).
Neal, R. M. Bayesian Learning for Neural Networks (Springer Science & Business Media, 2012).
Neal, R. M. in Bayesian Learning for Neural Networks Lecture Notes in Statistics Ch 2 (ed Nea, R. M.) 29–53 (Springer, 1996). This classic text highlights the connection between neural networks and Gaussian processes and the application of Bayesian approaches for fitting neural networks.
Williams, C. K. I. in Advances in Neural Information Processing Systems 295–301 (MIT Press, 1997).
MacKay David, J. C. A practical Bayesian framework for backprop networks. Neural. Comput. https://doi.org/10.1162/neco.1992.4.3.448 (1992).
Sun, S., Zhang, G., Shi, J. & Grosse, R. Functional variational Bayesian neural networks. Preprint at https://arxiv.org/abs/1903.05779 (2019).
Lakshminarayanan, B., Pritzel, A. & Blundell, C. Simple and scalable predictive uncertainty estimation using deep ensembles. Advances in Neural Information Processing Systems 30, 6402–6413 (2017).
Wilson, A. G. The case for Bayesian deep learning. Preprint at https://arxiv.org/abs/2001.10995 (2020).
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014).
Gal, Y. & Ghahramani, Z. in International Conference on Machine Learning 1050–1059 (JMLR, 2016).
Green, P. J. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82, 711–732 (1995).
Hoffman, M. D. & Gelman, A. The No-U-Turn Sampler: adaptively setting path lengths in Hamiltonian Monte Carlo. J. Mach. Learn. Res. 15, 1593–1623 (2014).
Liang, F. & Wong, W. H. Evolutionary Monte Carlo: applications to Cp model sampling and change point problem. Stat. Sinica 317-342 (2000).
Liu, J. S. & Chen, R. Sequential Monte Carlo methods for dynamic systems. J. Am. Stat. Assoc. 93, 1032–1044 (1998).
Sisson, S., Fan, Y. & Beaumont, M. Handbook of Approximate Bayesian Computation (Chapman and Hall/CRC 2018).
Rue, H., Martino, S. & Chopin, N. Approximate Bayesian inference for latent Gaussian models by using integrated nested Laplace approximations. J. R. Stat. Soc. Series B 71, 319–392 (2009).
Lunn, D. J., Thomas, A., Best, N. & Spiegelhalter, D. WinBUGS — a Bayesian modelling framework: concepts, structure, and extensibility. Stat. Comput. 10, 325–337 (2000).
Ntzoufras, I. Bayesian Modeling Using WinBUGS Vol. 698 (Wiley, 2011).
Lunn, D. J., Thomas, A., Best, N. & Spiegelhalter, D. WinBUGS — a Bayesian modelling framework: concepts, structure, and extensibility. Stat. Comput. 10, 325–337 (2000). This paper provides an early user-friendly and freely available black-box MCMC sampler, opening up Bayesian inference to the wider scientific community.
Spiegelhalter, D., Thomas, A., Best, N. & Lunn, D. OpenBUGS User Manual version 3.2.3. Openbugs http://www.openbugs.net/w/Manuals?action=AttachFile&do=view&target=OpenBUGS_Manual.pdf (2014).
Plummer, M. JAGS: a program for analysis of Bayesian graphical models using Gibbs sampling. Proc. 3rd International Workshop on Distributed Statistical Computing 124, 1–10 (2003).
Plummer, M. rjags: Bayesian graphical models using MCMC. R package version, 4(6) (2016).
Salvatier, J., Wiecki, T. V. & Fonnesbeck, C. Probabilistic programming in Python using PyMC3. PeerJ Comput. Sci. 2, e55 (2016).
de Valpine, P. et al. Programming with models: writing statistical algorithms for general model structures with NIMBLE. J. Comput. Graph. Stat.s 26, 403–413 (2017).
Dillon, J. V. et al. Tensorflow distributions. Preprint at https://arxiv.org/abs/1711.10604 (2017).
Keydana, S. tfprobability: R interface to TensorFlow probability. github https://rstudio.github.io/tfprobability/index.html (2020).
Bingham, E. et al. Pyro: deep universal probabilistic programming. J. Mach. Learn. Res. 20, 973–978 (2019).
Bezanson, J., Karpinski, S., Shah, V. B. & Edelman, A. Julia: a fast dynamic language for technical computing. Preprint at https://arxiv.org/abs/1209.5145 (2012).
Ge, H., Xu, K. & Ghahramani, Z. Turing: a language for flexible probabilistic inference. Proceedings of Machine Learning Research 84, 1682–1690 (2018).
Smith, B. J. et al. brian-j-smith/Mamba.jl: v0.12.4. Zenodo https://doi.org/10.5281/zenodo.3740216 (2020).
JASP Team. JASP (version 0.14) [computer software] (2020).
Lindgren, F. & Rue, H. Bayesian spatial modelling with R-INLA. J. Stat. Soft. 63, 1–25 (2015).
Vanhatalo, J. et al. GPstuff: Bayesian modeling with Gaussian processes. J. Mach. Learn. Res. 14, 1175–1179 (2013).
Blaxter, L. How to Research (McGraw-Hill Education, 2010).
Neuman, W. L. Understanding Research (Pearson, 2016).
Betancourt, M. Towards a principled Bayesian workflow. github https://betanalpha.github.io/assets/case_studies/principled_bayesian_workflow.html (2020).
Veen, D. & van de Schoot, R. Posterior predictive checks for the Premier League. OSF https://doi.org/10.17605/OSF.IO/7YRUD (2020).
Kramer, B. & Bosman, J. Summerschool open science and scholarship 2019 — Utrecht University. ZENODO https://doi.org/10.5281/ZENODO.3925004 (2020).
Rényi, A. On a new axiomatic theory of probability. Acta Math. Hung. 6, 285–335 (1955).
Lesaffre, E. & Lawson, A. B. Bayesian Biostatistics (Wiley, 2012).
Hoijtink, H., Beland, S. & Vermeulen, J. A. Cognitive diagnostic assessment via Bayesian evaluation of informative diagnostic hypotheses. Psychol Methods 19, 21–38 (2014).
Acknowledgements
R.v.d.S. was supported by grant NWO-VIDI-452-14-006 from the Netherlands Organization for Scientific Research. R.K. was supported by Leverhulme research fellowship grant reference RF-2019-299 and by The Alan Turing Institute under the EPSRC grant EP/N510129/1. K.M. was supported by a UK Engineering and Physical Sciences Research Council Doctoral Studentship. C.Y. is supported by a UK Medical Research Council Research Grant (Ref. MR/P02646X/1) and by The Alan Turing Institute under the EPSRC grant EP/N510129/1
Author information
Authors and Affiliations
Contributions
Introduction (R.v.d.S.); Experimentation (S.D., D.V., R.v.d.S. and J.W.); Results (R.K., M.G.T., M.V., D.V., K.M., C.Y. and R.v.d.S.); Applications (S.D., R.K., K.M. and C.Y.); Reproducibility and data deposition (B.K., D.V., S.D. and R.v.d.S.); Limitations and optimizations (A.G.); Outlook (K.M. and C.Y.); Overview of the Primer (R.v.d.S.).
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Methods Primers thanks D. Ashby, J. Doll, D. Dunson, F. Feinberg, J. Liu, B. Rosenbaum and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Dryad: https://datadryad.org/
Registry of Research Data Repositories: https://www.re3data.org/
Scientific Data list of repositories: https://www.nature.com/sdata/policies/repositories
Zenodo: https://zenodo.org/
Glossary
- Prior distribution
-
Beliefs held by researchers about the parameters in a statistical model before seeing the data, expressed as probability distributions.
- Likelihood function
-
The conditional probability distribution of the given parameters of the data, defined up to a constant.
- Posterior distribution
-
A way to summarize one’s updated knowledge, balancing prior knowledge with observed data.
- Informativeness
-
Priors can have different levels of informativeness and can be anywhere on a continuum from complete uncertainty to relative certainty, but we distinguish between diffuse, weakly and informative priors.
- Hyperparameters
-
Parameters that define the prior distribution, such as mean and variance for a normal prior.
- Prior elicitation
-
The process by which background information is translated into a suitable prior distribution.
- Informative prior
-
A reflection of a high degree of certainty or knowledge surrounding the population parameters. Hyperparameters are specified to express particular information reflecting a greater degree of certainty about the model parameters being estimated.
- Weakly informative prior
-
A prior incorporating some information about the population parameter but that is less certain than an informative prior.
- Diffuse priors
-
Reflections of complete uncertainty about population parameters.
- Improper priors
-
Prior distributions that integrate to infinity.
- Prior predictive checking
-
The process of checking whether the priors make sense by generating data according to the prior in order to assess whether the results are within the plausible parameter space.
- Prior predictive distribution
-
All possible samples that could occur if the model is true based on the priors.
- Kernel density estimation
-
A non-parametric approach used to estimate a probability density function for the observed data.
- Prior predictive p-value
-
An estimate to indicate how unlikely the observed data are to be generated by the model based on the prior predictive distribution
- Bayes factor
-
The ratio of the posterior odds to the prior odds of two competing hypotheses, also calculated as the ratio of the marginal likelihoods under the two hypotheses. It can be used, for example, to compare candidate models, where each model would correspond to a hypothesis.
- Credible interval
-
An interval that contains a parameter with a specified probability. The bounds of the interval are the upper and lower percentiles of the parameter’s posterior distribution. For example, a 95% credible interval has the upper and lower 2.5% percentiles of the posterior distribution as its bounds.
- Closed form
-
A mathematical expression that can be written using a finite number of standard operations.
- Marginal posterior distribution
-
Probability distribution of a parameter or subset of parameters within the posterior distribution, irrespective of the values of other model parameters. It is obtained by integrating out the other model parameters from the joint posterior distribution.
- Markov chain Monte Carlo
-
(MCMC). A method to indirectly obtain inference on the posterior distribution by simulation. The Markov chain is constructed such that its corresponding stationary distribution is the posterior distribution of interest. Once the chain has reached the stationary distribution, realizations can be regarded as a dependent set of sampled parameter values from the posterior distribution. These sampled parameter values can then be used to obtain empirical estimates of the posterior distribution, and associated summary statistics of interest, using Monte Carlo integration.
- Markov chain
-
An iterative process whereby the values of the Markov chain at time t + 1 are only dependent on the values of the chain at time t.
- Monte Carlo
-
A stochastic algorithm for approximating integrals using the simulation of random numbers from a given distribution. In particular, for sampled values from a distribution, the associated empirical value of a given statistic is an estimate of the corresponding summary statistic of the distribution.
- Transition kernel
-
The updating procedure of the parameter values within a Markov chain.
- Auxiliary variables
-
Additional variables entered in a model such that the joint distribution is available in closed form and quick to evaluate.
- Trace plots
-
Plots describing the posterior parameter value at each iteration of the Markov chain (on the y axis) against the iteration number (on the x axis).
- \(\hat{R}\) statistic
-
The ratio of within-chain and between-chain variability. Values close to one for all parameters and quantities of interest suggest the Markov chain Monte Carlo algorithm has sufficiently converged to the stationary distribution.
- Variational inference
-
A technique to build approximations to the true Bayesian posterior distribution using combinations of simpler distributions whose parameters are optimized to make the approximation as close as possible to the actual posterior.
- Approximating distribution
-
In the context of posterior inference, replacing a potentially complicated posterior distribution with a simpler distribution that is easy to evaluate and sample from. For example, in variational inference, it is common to approximate the true posterior with a Gaussian distribution.
- Stochastic gradient descent
-
An algorithm that uses a randomly chosen subset of data points to estimate the gradient of a loss function with respect to parameters, providing computational savings in optimization problems involving many data points.
- Multicollinearity
-
A situation that arises in a regression model when a predictor can be linearly predicted with high accuracy from the other predictors in the model. This causes numerical instability in the estimation of parameters.
- Shrinkage priors
-
Prior distributions for a parameter that shrink its posterior estimate towards a particular value.
- Sparsity
-
A situation where most parameter values are zero and only a few are non-zero.
- Spike-and-slab prior
-
A shrinkage prior distribution used for variable selection specified as a mixture of two distributions, one peaked around zero (spike) and the other with a large variance (slab).
- Continuous shrinkage prior
-
A unimodal prior distribution for a parameter that promotes shrinkage of its posterior estimate towards zero.
- Global–local shrinkage prior
-
A continuous shrinkage prior distribution characterized by a high concentration around zero to shrink small parameter values to zero and heavy tails to prevent excessive shrinkage of large parameter values.
- Horseshoe prior
-
An example of a global–local shrinkage prior for variable selection that uses a half-Cauchy scale mixture of normal distributions.
- Autoencoder
-
A particular type of multilayer neural network used for unsupervised learning consisting of two components: an encoder and a decoder. The encoder compresses the input information into low-dimensional summaries of the inputs. The decoder takes these summaries and attempts to recreate the inputs from these. By training the encoder and decoder simultaneously, the hope is that the autoencoder learns low-dimensional, but highly informative, representations of the data.
- Split-\(\hat{R}\)
-
To detect non-stationarity within individual Markov chain Monte Carlo chains (for example, if the first part shows gradually increasing values whereas the second part involves gradually decreasing values), each chain is split into two parts for which the \(\hat{R}\) statistic is computed and compared.
- Amortization
-
A technique used in variational inference to reduce the number of free parameters to be estimated in a variational posterior approximation by replacing the free parameters with a trainable prediction function that can instead predict the values of these parameters.
Rights and permissions
About this article
Cite this article
van de Schoot, R., Depaoli, S., King, R. et al. Bayesian statistics and modelling. Nat Rev Methods Primers 1, 1 (2021). https://doi.org/10.1038/s43586-020-00001-2
Accepted:
Published:
DOI: https://doi.org/10.1038/s43586-020-00001-2
This article is cited by
-
“High-throughput screening of catalytically active inclusion bodies using laboratory automation and Bayesian optimization”
Microbial Cell Factories (2024)
-
Genomic data integration tutorial, a plant case study
BMC Genomics (2024)
-
Rapid reaction optimization by robust and economical quantitative benchtop 19F NMR spectroscopy
Nature Protocols (2024)
-
X-ray diffraction under grazing incidence conditions
Nature Reviews Methods Primers (2024)
-
Impact of genetic predisposition to late-onset neurodegenerative diseases on early life outcomes and brain structure
Translational Psychiatry (2024)