Abstract
Atmospheric aerosol and the cloud droplets and ice crystals that grow on them remain major sources of uncertainty in global climate models. A subset of aerosol, ice nucleating particles, catalyze the freezing of water droplets at temperatures warmer than −38 °C. Here we show that RuBisCO, one of the most abundant proteins in plants and phytoplankton, is one of the most efficient known immersion ice nucleating particles with a mean freezing temperature of −7.9 ± 0.3 °C. Further, we demonstrate RuBisCO is present in ambient continental aerosol where it can serve as an ice nucleating particle. Other biogenic molecules act as immersion ice nucleating particles, in the range of −19 to −26 °C. In addition, our results indicate heat denaturation is not a universal indicator of the proteinaceous origin of ice nucleating particles, suggesting current studies may fail to accurately quantify biological ice nucleating particle concentrations and their global importance.
Similar content being viewed by others
Introduction
Atmospheric ice nucleation impacts global climate and hydrological cycling through cloud formation, lifetime, albedo, and precipitation efficiency1. Ice crystal formation from pure water droplets in the atmosphere occurs at temperatures below −38 °C through homogeneous freezing2. In the presence of ice nucleating particles (INPs), heterogeneous freezing is catalyzed at warmer temperatures and lower supersaturations2,3. The most common mode of heterogeneous ice nucleation in the troposphere is immersion freezing, which occurs when an INP immersed inside a water droplet initiates the freezing process2,4. INPs are rare in the environment, representing only 1 in 105 (or less) aerosol particles5, but represent significant influence on tropospheric clouds given that the mass of ice is greater than liquid water in clouds, even in the tropics1. The sources, composition, and properties of INPs, especially biological ones, are not well understood, leading to climate model inaccuracies in representing ice-containing clouds and uncertainties in cloud phase partitioning and radiative transfer1,6.
The ice nucleating properties of non-biological aerosols, including mineral dust7,8, anthropogenic soot and pollutants9,10 are better understood, though in the case of soot, ice nucleation temperatures vary due to the influences of atmospheric aging, degree of oxidation, and ice nucleation mechanism10,11,12,13. Biological sources of INPs include pollen14,15,16,17, phytoplankton18,19, Archaea20, Bacteria21,22, fungi23,24,25, and viruses26. Pollen from some tree species15 and fungi27 are highly effective ice nuclei, catalyzing freezing at temperatures > −10 °C. This recent attention to biological molecules is warranted, since some of the warmest identified ice nucleators are of biological origin2,28,29.
Specific physical characteristics have been proposed to increase ice nucleation efficiency, including hexagonal crystalline structures akin to hexagonal ice and increased particle surface area2,30. Increased viscosity has been shown to reduce immersion mode ice nucleation2,31, but increase contact mode ice nucleation for organic compounds32. Chemical functional groups on the surface of an INP, such as hydroxyl and amine groups, are hypothesized to initiate freezing by aligning water molecules in a similar structure to ice crystals33.
Most previous work on biogenic INPs has focused on complex mixtures of organic matter, often composed of both whole microorganisms and non-living organic matter19,34,35,36. Ice nucleation measurements made on a single taxon of microorganism grown in pure cultures20,21,36,37,38 represent simpler systems. However, even samples containing one microorganism are incredibly complex. For example, the diatom Thalassiosira pseudonana used in ice nucleation experiments38 contains genes encoding 11,242 different proteins39. The complexity of even simple living organisms means that it is challenging to work out which compounds, or even class of compound, affect ice nucleation. Despite this complexity, biogenic ice nucleation has been modeled as a single INP type with homogeneous properties40.
Few previous studies have investigated individual compounds as biogenic INPs. Cellulose, a major component of cell walls in plants and some microorganisms, catalyzes ice via immersion mode nucleation41,42 as does lignin, another major component of cell walls43. Polysaccharides (amylopectin and agarose) and an amino acid (aspartic acid) had similar deposition nucleation efficiencies to aerosolized Prochlorococcus, an abundant marine cyanobacterium, suggesting these compounds may be important components of the INP population in the cirrus cloud regime44. Pseudomonas syringae, a terrestrial bacterium, produces a protein which is the most efficient known biological INP, with an onset freezing temperature of −1.8 °C and completely activated by −12 °C45.
In this study, we take a reductionist approach by focusing on representative globally abundant biogenic molecules (amino acids, proteins, and nucleic acids) and identifying their ice nucleation potential. Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) was selected as an example of a complex protein. It is one of the most abundant proteins on Earth and is found ubiquitously in marine and terrestrial photosynthetic organisms46,47,48. RuBisCO (~550 kDa)49 comprises up to 65% of total soluble protein in leaves and 2 to 23% of the total protein in phytoplankton46,50. Similarly, nucleic acids were included as they are complex biopolymers whose ice nucleation potential had not been measured. Our results show that all these organic compounds are immersion INPs, catalyzing the freezing of water droplets at temperatures significantly warmer than pure water procedural blanks.
Results and discussion
Ice nucleation activity of RuBisCO compared with other abundant biomolecules
Ice nucleation measurements were conducted in immersion mode using our custom-built ice microscope apparatus using a droplet volume of 2 µL with a minimum of 5 independent replicates and up to 25 freeze-thaw cycles for each replicate10,51. Compounds were assigned the following descriptive terms based on their mean immersion freezing temperature: −32 to −25 °C were described as ‘weakly effective’, −25 to −20 °C as ‘moderately effective’, −20 to −10 °C were described as ‘effective’ and > −10 °C as ‘highly effective’.
All living organisms contain proteins and polypeptides, which have a primary structure composed from sequences of amino acids. For reference, a structural representation of RuBisCO is shown in Supplementary Fig. 1. The number of different proteins on Earth is on the order of 100 s of millions, precluding untargeted screening for INPs52. However, all proteins on Earth are built from only 20 different amino acids. In this study, we evaluated representative individual amino acids as potential INPs. The mean freezing temperatures and number of replicates are summarized in Table 1 and Fig. 1. The mean nucleation temperatures of threonine (−26.2 ± 2.7 °C; weakly effective) and aspartic acid (−19.1 ± 2.0 °C; effective) represented the coolest and warmest temperatures, respectively. Glutathione (0.3 kDa) is a tripeptide (composed of glutamic acid, cysteine, and glycine) found in cells at concentrations of 0.1–10 mM53,54. The relative structural complexity of glutathione compared with individual amino acids (Supplementary Table 1), did not affect its ice nucleation properties (Fig. 1). It was a moderately effective INP, with a mean nucleation temperature of −21.8 ± 2.2 °C.
Mean nucleation temperatures of amino acids (blue), a peptide (red), nucleic acids (green), and protein (enzymes) (orange) with error bars representing the pooled standard deviation (n = 41–147; Table 1). Concentrations were measured at 5 mg mL−1 (circle), 0.5 mg mL−1 (diamond), and 5 × 10−4 mg mL−1 (square) with the lower concentration of cysteine (0.5 mg mL−1) shown in light blue and the lower concentration of DNA (5 × 10−4 mg mL−1) shown in light green. The black dashed line represents the mean freezing temperature of the procedural blank (HPLC water) in our apparatus and the pooled standard deviation (gray shaded region; n = 127).
Heterogeneous ice nucleation is a stochastic process which occurs over a range of temperatures. To visualize the temperature range of freezing events, fraction frozen was plotted for each of the amino acids and peptide in this study in Fig. 2a. For comparison, a procedural blank (HPLC water which was treated to all the same handling procedures as the samples) is also included. As can be seen in the figure, all the amino acids and peptides nucleated ice at temperatures well above the procedural blank, indicating that all serve as immersion mode INPs. This result was confirmed statistically using a non-parametric analysis of variance (Kruskall–Wallis test), followed by post-hoc analysis (Wilcoxon method) that showed the procedural blank froze at a significantly different temperature than all of the compounds (Supplementary Table 2). The ice nucleation active site density, or number of active sites per mg of organic compounds, is plotted in Fig. 2b. All samples had a concentration of 5 mg mL−1 except cysteine, which was measured at both 5 mg mL−1 (dotted green line) and 0.5 mg mL−1 (solid green line). There was no significant difference in the median nucleation temperature of cysteine at the two concentrations (Fig. 1, Supplementary Table 2). In addition, glutathione was measured in the native state (solid pink line) and heat denatured state (dashed pink line). As can be seen in Fig. 2b, the number density of ice nucleation active sites at a single temperature, for example −20 °C, spans >3 orders of magnitude between the different amino acids.
a Fraction of droplets frozen as a function of temperature for each organic compound. b Cumulative number of active sites per mass in mg of each compound as a function of temperature, nm(T), (n = 41–115; Table 1). The black line in a represents the fraction of droplets frozen as a function of temperature for the procedural blank (HPLC water; n = 127). Each unique compound is represented by a different color with glycine (red), threonine (blue), cysteine (green), aspartic acid (orange), lysine (purple), tryptophan (light blue), and glutathione (pink). All samples have a concentration of 5 mg mL−1 except cysteine which was measured at 5 mg mL−1 (dotted line) and 0.5 mg mL−1 (solid line). Glutathione (0.5 mg mL−1) was measured in the native state (solid line) and heat denatured state (dashed line).
The enzyme RuBisCO is a highly effective INP (Fig. 1), with the warmest observed mean freezing temperature in this study (−7.9 ± 0.3 °C), including the highest fraction frozen with initial freezing at −6.8 °C and complete freezing at −9 °C (Fig. 3a). In contrast, the enzyme alkaline phosphatase (140 kDa) is a moderately effective INP with a mean nucleation temperature of −20.4 ± 1.3 °C (Fig. 1). The ice nucleation active site density increases with decreasing temperature to a maximum of 4.7 × 103 INPs mg−1 alkaline phosphatase at −25.4 °C and colder, compared with a maximum of 4.8 × 103 INPs mg−1 RuBisCO at −8.8 °C and colder (Fig. 3b). Our data shows that RuBisCO nucleates ice at warm temperatures approaching those reported for the InaZ protein from Pseudomonas syringeae21. InaZ is used as an ice nucleation protein by P. syringeae, whereas RuBisCO is an essential enzyme in photosynthetic carbon fixation. Therefore, the ice nucleation properties of RuBisCO cannot be attributed to its primary biological function. This confirms previous work showing that large and complex proteins are highly effective immersion ice nuclei, irrespective of function55.
a Fraction of droplets frozen as a function of temperature for each organic compound. b Cumulative number of active sites per mass in mg of each compound as a function of temperature (nm(T)), (n = 46–147; Table 1). The black line in a represents the fraction of droplets frozen as a function of temperature for the procedural blank (HPLC water; n = 127). Each unique compound is represented by a different color with DNA (red), RNA (blue), alkaline phosphatase (green), and ribulose-1,5-carboxylase/oxygenase (RuBisCO; purple). All samples were measured at 0.5 mg mL−1 except DNA. DNA from laboratory grown Synechococcus elongatus was measured at 5.0 × 10−4 mg mL−1 (dotted line), while DNA from herring sperm at 0.5 mg mL−1 (solid line). The two enzymes, alkaline phosphatase and RuBisCO, were measured in the native state (solid line) and heat denatured state (dashed line).
Like proteins, nucleic acids are large biopolymers composed of sequences of subunits. Our results show that nucleic acids had a range of nucleation temperatures and were moderately effective or effective INPs (Figs. 1 and 3). Freezing onset in RNA occurred at −12.6 °C, with complete freezing at −26.8 °C, which was a larger range of temperatures than observed for DNA (Fig. 3a). DNA was found to be enriched by a factor as high as 30,000 in artificially generated sea spray aerosol using seawater from the North Atlantic56. Given the ubiquity of nucleic acids in living systems and the detection of nucleic acids in aerosol samples57,58,59, nucleic acids could contribute to atmospheric INPs.
In Fig. 3, fraction frozen (Fig. 3a) and ice nucleation active site density (Fig. 3b) are shown for the nucleic acid and enzyme samples. The active site density as a function of temperature is controlled by freezing efficiency as well as concentration. Due to their lower solubility, nucleic acids and enzymes were measured at 0.5 mg mL−1, rather than the 5 mg mL−1 concentration used for amino acids. An exception was DNA (Fig. 3), which was measured from two sources: herring sperm at 0.5 mg mL−1 (solid red line) and Synechococcus elongatus at 5 × 10−4 mg mL−1 (dotted red line). This variation in sample concentration was necessary due to the limited amount of Synechococcus elongatus DNA that was recovered from a laboratory grown culture. As can be seen in Fig. 3, the lower concentration DNA (5 × 10−4 mg mL−1) exhibited a fraction frozen active a few degrees colder than the 0.5 mg mL−1. However, in terms of the number of active sites per mg of material, the lower concentration DNA provided a much higher ice nucleation active sites per unit mass of organic sample material, nm (Eq. 2).
To address the atmospheric relevance of this work, the efficiency of the materials chosen here were compared to other biological INP. At −20 °C, the majority of samples in this study have nm in the range of ~100–1000 mg−1 (Figs. 2b and 3b). Exceptions are the DNA extracted from Synechococcus elongatus with a higher nm of 6.4 × 105 mg−1 at −20 °C, and RuBisCO whose maximin nm of 4.8 × 103 mg−1was already achieved by −9 °C. Comparing to previous studies, we see that estimates of nm (−20 °C) for humic like substances (HULIS) range from between 213 and 8.7 × 104 per mg at −20 °C60, in broad agreement with the results for the majority of our samples at −20 °C. There is a wide spread in nm values according to measurements for pollen, fungi, ferritin, and apoferritin depending on temperature and material6. Broadly speaking, on a gram per gram basis, RuBisCO is approximately as effective at nucleating ice at warm temperatures above −10 °C as other biological materials are at colder temperatures between −10 and −25 °C.
Proteins are composed of linear sequences of amino acid residues bonded covalently (primary structure), which occupy three-dimensional space to form the secondary and tertiary structure, which is stabilized by non-covalent interactions61. Large proteins, including RuBisCO, have a quaternary structure as they are formed from multiple polypeptide subunits (Supplementary Fig. 1). In theoretical studies, both the sequence of amino acids (primary structure) and how those resulting polypeptides are orientated (secondary, tertiary, and quaternary structure) contributes to heterogeneous freezing33,62,63. It has been proposed that repeating sequences of amino acids located on β-helical structures (secondary structure) serve as energetically favorable sites for ice-binding33,63. RuBisCO is rich in amino acids that are associated with ice nucleation (threonine (T), serine (S), tyrosine (Y) and glutamic acid (E) (Supplementary Fig. 2))62,63. We suspect the large size64 and complex protein folding with β-helical structures at the surface of RuBisCO contribute to the ordering of water molecules and efficient ice nucleation. Recent work proposed that ice nucleation activity is a common feature of proteins and may arise through defective folding or aggregation into structures that are active ice nuclei55. The formation of aggregates of protein molecules may be affected by protein concentration in the water; apoferritin and ferritin were less effective as INPs when solutions were diluted (e.g., from 0.34 to 3.4 × 10−4 mg mL−1), which may be due to disaggregation and even the disassembly of individual proteins into subunits55. We measured the ice nucleation activity of RuBisCO at 0.5 mg mL−1 and therefore do not know how dilution would affect its ice nucleation.
Heat denaturation treatments of proteins in biogenic aerosol is not always effective
In over 40 studies since 1972, denaturing and oxidation treatments have been used as a tool to determine whether INPs have a biogenic origin. INP activity and nucleation temperature have shown to be reduced upon heat in many studies, as summarized in Supplementary Table 3. Heating (generally 90 to 100 °C) is used to denature proteins in aerosol samples, whereas hydrogen peroxide is used to remove organic matter by oxidation. Denaturation is defined as disruption in the secondary, tertiary, and quaternary structure which leads to a reduction or loss of function65. For example, RuBisCO unfolds, disrupting its secondary and tertiary structure, with a complete loss of enzymatic activity at temperatures over 65 °C66. Here, heat treatment (15 min at 95 °C in water) of RuBisCO resulted in a significant decrease in ice nucleation temperature, from a mean of −7.9 ± 0.3 °C to −21.4 ± 1.3 °C (Fig. 4), suggesting that the secondary, tertiary, or quaternary protein structure was essential to its ice nucleation activity at >−10 °C. While heat denaturation significantly decreased the median nucleation temperature compared with the native state (Table 1), it did not eliminate INP activity. Denatured RuBisCO remained a moderately effective INP, with complete freezing observed below −25.2 °C (Fig. 3a). At temperatures colder than −25 °C, denatured RuBisCO had similar active site density (4.8 × 103 INPs mg−1) as native RuBisCO at −8.8 °C (4.6 × 103 INPs mg−1) (Fig. 3b). Identical heat treatment did not reduce the nucleation temperature of glutathione and alkaline phosphatase (Figs. 2 and 3). As a simple tripeptide, glutathione does not have a secondary and tertiary structure to disrupt with heating. Alkaline phosphatase has complex secondary and tertiary structure, yet heating caused a relatively small decrease in mean nucleation temperature from −20.4 ± 1.3 °C to −22.6 ± 1.3 °C (Fig. 3; Table 1). Alkaline phosphatase can refold and resume enzymatic activity after denaturation by heat treatment67 and upon cooling to temperatures <40 °C68,69. It is also possible that while heating is effective at disrupting protein shapes and eliminating enzyme activity, it does not eliminate ice nucleation activity as the proteins are twisted into new shapes that have ice nucleation sites, similar to the hypothesis that defects and aggregation of large proteins are a source of ice nucleation sites55. Our work shows that individual amino acids and a simple oligopeptide (glutathione) are also ice nuclei. Collectively, these findings suggest that ice nucleation is not dependent on specific secondary, tertiary and/or quaternary structures, or even specific amino acid motifs in the primary structure of proteins. Further work is needed to determine whether interactions, such as hydrogen bonding, between free amino acids or oligopetides are important in determining ice nucleation properties. This could be approached by manipulating concentration and pH of the solutions and measuring ice nucleation55.
Mean immersion nucleation temperatures of native (blue) and denatured (red) peptide (glutathione) and enzymes (alkaline phosphatase and ribulose-1,5-carboxylase/oxygenase (RuBisCO)) with error bars representing the pooled standard deviation (n = 46-98). All compounds were measured at a concentration of 0.5 mg mL−1. The black dashed line represents the mean freezing temperature of procedural blank (HPLC water) and pooled standard deviation (gray shaded region; n = 127).
Source apportionment is necessary to determine the relative contribution of different categories of aerosol to atmospheric INPs and for realistic parametrization of biogenic ice nucleation in models6,70. Recent work55,71 including our own data, shows that commonly applied ‘wet’ heating (warming solutions or suspensions of potential INP to 90–100 °C; Supplementary Table 3) is inadequate for determining biogenic sources of INPs as the effects of heating are much more complex than previously realized. Our results demonstrate that heat treatment cannot be used in isolation to determine whether immersion mode INPs contain proteinaceous material. Some polypeptides and proteins are resistant to heating or are capable of reverting to the native state after denaturation72. In addition, free amino acids are moderately effective INPs. By ignoring this complexity, current field assessments of biogenic INPs may misrepresent the range of temperature behaviors in the immersion mode for biogenic INPs. Studies that attribute the remaining ice nucleation activity after heat treatment to non-biological sources will greatly underestimate the total biological INP concentration. Our results show that heat denaturation is likely to underestimate the contribution of proteins and other biomolecules in the INP population, particularly ice nucleation at colder temperatures (<−20 °C), where it is assumed that non-biogenic INPs dominate, such as mineral dust6. A previous study also used dry heating of potential INPs at higher temperatures (250 °C for 4 h)71. Although effective at deactivating biogenic INPs, dry heat also deactivated the most significant ice-nucleating mineral (K-feldspar) in atmospheric dust. Unlike our laboratory experiments, field samples generally contain a mixture of INPs from both biogenic and non-biological sources, making interpretation of loss of INP activity on heating, and source apportionment, extremely challenging71.
RuBisCO detected in atmospheric aerosol
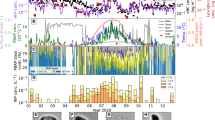
RuBisCO has not been observed in atmospheric aerosol prior to this study. To confirm its atmospheric presence, high-volume samplers were used to collect atmospheric aerosol particles (0.15 to 12 µm aerodynamic diameter) from a position 47 m above ground level on a rooftop in College Station, Texas, USA (30°37’4”N, 96°20’ 11”W) (Methods). Atmospheric RuBisCO concentrations were determined using an enzyme-linked immunosorbent assay (ELISA). RuBisCO was present in atmospheric aerosol, with concentrations between 4.3 × 10−10 and 1.9 × 10−9 mg L−1 air (Fig. 5a). The measured atmospheric aerosol concentrations were above the detection limit of 1.9 × 10−10mg L−1 of air. The limit of detection was determined based on the volume of air filtered and data provided by the manufacturer of the ELISA kit. The variation between analyses of material from the same filter may indicate that RuBisCO was not homogeneously distributed throughout the sample that may contribute a significant number of highly effective INPs active at warm temperatures. While only collected at one continental site, these data demonstrate that RuBisCO, a ubiquitous component of plant life, is present in the ambient atmosphere. HYSPLIT back trajectories were used to estimate where the sampled airmasses had come from over the 7 days prior to the end of the sampling period. Airmasses were predicted to have passed over both land (Great Plains of North America) and ocean (Gulf of Mexico and North Atlantic) (Supplementary Fig. 3).
a Mean concentration and standard deviation of each sample replicate of ribulose-1,5-carboxylase/oxygenase (RuBisCO) in atmospheric aerosol sampled in College Station (Texas, United States; 30.62°N, 96.34°S) during September 2021 on three separate days. Replicate measurements were made from a single aerosol sample collected over 48 h; n = 2 on 09/13 and n = 3 for 09/19 and 09/21. Dates represent the end of each 48 h sampling period. The detection limit for quantifying RuBisCO in the volume of air sampled was 1.9 × 10−10mg L−1 of air. b Estimated number of ice nucleation active sites from RuBisCO per L of air in atmospheric aerosol. The number of active sites was calculated using laboratory ice nucleation measurements and the measured mean atmospheric aerosol concentrations of RuBisCO on September 11–13, 2021 (green), September 17–19, 2021 (blue), and September 19–21, 2021 (red).
Ambient number of active sites per liter of air (nsite) was estimated accordingly to Eq. 4 (Fig. 5b), using the laboratory measurements of number of active sites per mass of RuBisCO combined with the ambient RuBisCO concentrations. At −9 °C, nsite ranged from 2.0 × 10−6 to 9.1 × 10−6 ice nucleation active sites L−1 air, which represents a significant number of highly effective INPs active at warm temperatures. At the mean freezing temperature of RuBisCO (−7.9 °C), nsite ranges from 5.6 × 10−7 to 1.7 × 10−6 ice nucleation active sites L−1 air. Similar to our approach with RuBisCO, combined laboratory measurements of nsite with ambient measurements of humic like substances (HULIS) were used to estimate the active site density L−1 air60. Compared to ambient measured values of INPs in the aerosol population and in precipitation, they concluded that HULIS may be a significant source of INPs at temperatures as warm as ~−10 °C60. Overall, the number of ice nucleation active sites we observed for RuBisCO are lower than those of HULIS. However, there is a strong temperature dependence to nsite concentrations. At temperatures of −9 °C and warmer, RuBisCO may be a significant fraction of INP, since nsite is comparable to the total INP concentration observed in precipitation in the continental United States on low INP days73. Additional measurements are needed to assess geographical and seasonal variation in RuBisCO. Broadly speaking, observed RuBisCO ice active fractions may represent a significant concentration of ice nucleation active sites, particularly when other sources of warm temperature INPs are not present. Our measurements demonstrate the presence of ambient RuBisCO in the atmospheric and provide motivation for further field measurements to assess variations in RuBisCO contributions to INP concentrations according to geographic location and time.
Implications for understanding biogenic ice nucleation
Our data show that RuBisCO is one of the most effective known biogenic INPs, initiating immersion freezing at temperatures as warm as −6.8 °C, ~31 °C warmer than the homogeneous freezing of water droplets in the atmosphere. RuBisCO is one of the most abundant proteins on Earth (~0.7 Pg; where 1 Pg = 1 × 1015 g), with a global distribution in both terrestrial plants and marine phytoplankton46,47,48. We demonstrated that RuBisCO is present in the atmosphere, though further work is needed to determine its atmospheric significance. RuBisCO has been measured in the Pacific Ocean in µg L−1 concentrations at depths down to 4000 m74. Measurements on filtered water collected well below the depths where phytoplankton are active (generally <200 m) indicate that RuBisCO is found in the environment outside of living organisms and is relatively stable, at least in the ocean. Processes that break open phytoplankton cells, such as sloppy feeding by zooplankton75,76 and viral lysis77,78, provide mechanisms that release RuBisCO and other cell contents into the water. It is well established that dissolved organic matter is a component of sea spray aerosol79,80,81. It is harder to envision how RuBisCO from terrestrial plant leaves enters the atmosphere. However, microalgae are also a significant component of the terrestrial biosphere and are detected in the atmosphere18,57,82,83. The estimated average global concentration of microalgae in surface soils is (5.5 ± 3.4) × 106 cells per gram84. Once in the atmosphere, cell lysis caused by osmotic shock on immersion in cloud water85 provides another potential mechanism to release RuBisCO into the environment.
Potentially, atmospheric RuBisCO widens the range of conditions under which ice occurs in clouds, with global significance for the properties of both ice and mixed-phase clouds, and the Earth’s radiative budget and climate. There are unexplained episodic high freezing temperature INPs over marine regions70, and the highly ice-active enzyme RuBisCO may partially account for these high freezing temperatures. However, these are the first known measurements of RuBisCO in the atmosphere and further research is needed to constrain their atmospheric relevance from both terrestrial and marine environments. Overall, the importance of proteins as biogenic INPs is currently underestimated and requires further investigation. We suggest that large and complex protein molecules, specifically those which are abundant in ecosystems and globally distributed, warrant further investigation as potentially climatically significant INPs.
Further work is needed to incorporate biogenic aerosol into global climate models, particularly reducing uncertainties associated with cloud processes1,86. However, our results, considered with those from a previous study71, show that the response to heat denaturation is complex, and there are multiple problems with using heat treatments to determine the concentrations of INP which are of biological origin. Incomplete deactivation of biological INPs will lead to an underestimation of their contribution in the environment. In addition, the misclassification of heat sensitive mineral dust (i.e., quartz, plagioclase feldspars, and Arizona test dust) as biological INPs could also lead to an overestimation of biological INP concentration. Further work is needed to develop a method to assess the relative importance of these sources.
Methods
Sample preparation for ice nucleation experiments
Aqueous solutions were prepared at concentrations of 5 mg mL−1, 0.5 mg mL−1, and 5 × 10−4 mg mL−1 using ultrapure water (HPLC water, Sigma-Aldrich) and commercially available compounds, which were obtained from Sigma-Aldrich unless otherwise noted. Lower concentrations were used when it was not possible to make measurements at 5 mg mL−1, either due to solubility issues (i.e. enzymes and nucleic acids) or the available mass of sample (i.e. DNA from the globally abundant marine cyanobacterium Synechococcus elongatus). Ribulose-1,5-biphosphate carboxylase-oxygenase (RuBisCO) originated from spinach (Spinacia oleracea) and is representative of the most abundant type (Form I) on Earth, found in land plants and most phytoplankton groups87. Alkaline phosphatase originated from calf (Bos taurus) intestine, deoxyribonucleic acid (DNA) from herring (Clupea sp.) testes, and ribonucleic acid (RNA) from Escherichia coli (Invitrogen E. coli Total RNA, Fisher Scientific). The purchased RNA was further purified to remove the 1 mM sodium citrate buffer it was supplied in. The buffer was removed using isopropanol precipitation with 3 M sodium acetate (pH 6.0) and centrifuged at 12,000 x g for 30 min. The RNA was washed again with 70% ethanol and centrifuged at 12,000 x g for 30 min, followed by resuspension in ultrapure water. In addition to a commercially available source of DNA, total DNA was extracted from a batch culture of Synechococcus elongatus using FastDNA Spin kits (MP Biomedical) from filters88 and resuspended in ultrapure water. Nucleic acid was quantified using the Quantifluor dsDNA (double-stranded DNA) and RNA system (Promega). All aqueous solutions of chemical compounds were stored at −20 °C for up to 1 week after preparation for ice nucleation measurements.
The molecular formula and weight for nucleic acid (Supplementary Table 1) was estimated using the GC (guanine-cytosine) content of Synechococcus elongatus (60.67%)89 for DNA and Escherichia coli for RNA (50.8%)90. The percentages of the remaining nucleobases, adenine and thymine for DNA (39.33%) and adenine and uracil for RNA (49.2%), were calculated from the GC content. The sequence size of nucleic acid can vary; a fragment length of 1500 base pairs was used as an average length. The total number of carbons, hydrogen, oxygen, nitrogen, and phosphorus atoms were determined based on the nucleotides containing a nitrogenous base, sugar backbone, and phosphate group to calculate the molecular formula and weight.
Ice nucleation
Ice nucleation measurements were conducted with our well-established ice microscope technique19,32,51, consisting of a cooling stage (Linkam Scientific Instruments, LTS 120) mounted to an optical microscope (Olympus BX43F). This setup allows a series of freezing temperature data points from a single droplet using a previously described method51. Freeze-thaw cycles from +5 °C to −40 °C were maintained with a temperature accuracy of ±0.2 °C. A three-point temperature calibration was performed of the cooling stage to verify the accuracy of the recorded temperature for our experiments using n-dodecane, n-undecane, and n-decane32,51. These compounds were used based on their well-documented melting temperatures across a range of temperatures suitable for the cooling stage. The estimated error of the cooling stage was ±0.2 °C over the experimental temperature range.
Ice nucleation was measured in the immersion mode, in which a 2 μl droplet from aqueous solutions described above, was micro-pipetted inside the sealed cooling stage on aluminum foil and a silanized glass microscope slide for support. The slide was silanized by spraying a hydrophobic coating (Rain-X water repellent, ITW Global Brands) on the entire slide and completely dried before use. The temperature of the stage was controlled using a temperature controller (Linkam Scientific Instruments, T96) and a water circulation bath (VWR) circulating water within the cooling stage to allow cooling to −40 °C. The droplet was cooled at a rate of 1 °C min−1 to −40 °C using LabSpec 6.2 software (Horiba Scientific). Once the stage reached −40 °C, the droplet was warmed at a rate of 10 °C per min−1 to 5 °C where it remained at 5 °C for 1 min to ensure complete thawing. The freezing temperature was recorded using images taken every 0.2 °C at 5x magnification (Syncerity charge-coupled device (CCD), Horiba Scientific). The freeze-thaw cycle was repeated to record multiple ice nucleation events of the same sample. Droplet volume was maintained during freeze-thaw cycles using a constant flow of humidified nitrogen generated by combining humid nitrogen (0.01–0.05 L min−1) from a bubbler containing ultrapure water with dry nitrogen gas (0.6 L min−1) and passed through a mixing chamber before entering the cooling stage. Gas flow rates were controlled using mass flow controllers (Alicat Scientific) and the humidity and dew point monitored using a dew point hygrometer (EdgeTech DewPrime II). At least 5 independent replicates of each compound were maintained up to 25 freeze-thaw cycles, with a minimum of 3 freezing points for each replicate. Freezing points were not analyzed when the droplet had visually changed size or if condensation was present on the microscope slide on the droplet. After each experimental run, CCD images were analyzed frame-by-frame to determine the freezing temperature of each nucleation event, determined by the opacity of the droplet, where liquid droplets were transparent and frozen droplets were opaque. The fraction frozen and cumulative number of ice nucleation active sites per unit mass were calculated from the individual freezing data points3,91.
Ice nucleation calculations
In Figs. 2a and 3b above, the probability of freezing (P(T)) at a given temperature (T), or fraction frozen, was calculated as:
where Nf is the cumulative number of droplet freezing events at a given temperature or warmer, and No is the total number of freezing events for a sample compiled from all replicates91.
In Figs. 2b and 3b the cumulative number of ice nucleation active sites per unit mass of organic sample material (nm(T)) was calculated as:
where Vdroplet is the volume of one sample droplet (2 µL in this study) and Cm is the mass concentration of organic material in the sample droplet3,9,91.
To assess the atmospheric significance of these new findings, the potential contribution of RuBisCO to the total ice nucleation active sites per L of air at a terrestrial site was estimated based on concentration of ice nucleation active sites per unit mass of RuBisCO (nm of RuBisCO) measured in the laboratory (Eq. 2) and the mass concentration of RuBisCO (Cair in mg of RuBisCO per L of air) from the Davis Rotating-drum Universal-size-cut Monitoring (DRUM) impactor stages (as described below).
First, the ambient Cair was determined from measurements collected in 48-h periods:
where M is the total mass of RuBisCO sampled on the impactors during collection time (T) and F is the sample flowrate through the DRUM (L−1).
Next, the ambient number of active sites per liter of air (nsite) was estimated according to Eq. 4:
Heat denaturation of proteins
Aqueous solutions of the proteins (RuBisCO and alkaline phosphatase) and peptide (glutathione) were denatured by heat treatment, which has been used in many previous studies as summarized in Supplementary Table 3. The solutions were placed in 15 mL sterile polypropylene centrifuge tubes with screw caps (VWR), which were denatured by suspending them in a water bath at 95 °C for 15 min. Ice nucleation measurements were conducted immediately after heat treatment to determine the ice nucleation temperature of the denatured proteins and peptide. The nucleation temperatures of both the native and denatured states were determined at the same concentrations in aqueous solution (see the sample preparation section above).
RuBisCO sample collection and analysis
Ambient aerosol samples were collected on the campus of Texas A&M University in College Station, Texas USA (30° 37’4” N, 96° 20’ 11” W; ~250 m above sea level) from 11 September to 21 September 2021. The sampling site was 47 m above ground level on the flat roof of the Oceanography and Meteorology Building. A Davis Rotating-drum Universal-size-cut Monitoring (DRUM) impactor (DRUMAir DA400) collected aerosol particles (0.15–12 µm in diameter) from 25 L air min−1 for 48 h. Samples were collected on aluminum foil strips at a rate of 5 mm day−1 in each stage of the 4 stages of the impactor. The aluminum foil strips were combusted (500 °C for 6 h) to remove potential organic contamination prior to loading into the DRUM impactor sampler. Aerosol from all four stages (0.15–12 µm in diameter) were pooled and washed into a single 1.5 mL autoclaved microfuge tube using 500 µL ultrapure water. Samples were stored at −80 °C for up to 2 weeks.
RuBisCO was quantified using a RuBisCO enzyme-immunoassay (ELISA) kit (GenWay Biotech Inc.), measuring over the range 13.7–10,000 ng mL−1. Prior to analysis, the aerosol sample was sonicated (QSonica Q125) for 1 min using 5 s pulses, with the tube on ice, to prevent heat buildup. Sonicated samples were stored at 4 °C for 30 min, then centrifuged at 2660 x g for 10 min and the supernatant was collected for the RuBisCO ELISA. The RuBisCO ELISA kit uses standard sandwich enzyme-linked immune-sorbent assay technology. The samples were added to a 96-well plate pre-coated with an antibody and a second horseradish peroxidase (HRP) conjugated detector antibody specific for RuBisCO. The enzymatic reaction resulted in a color change and the optical density was determined by absorbance at 450 nm using an UV-Vis spectrophotometer (Tecan Spark).
Back trajectory models
Back trajectory models were used to estimate where the air sampled for RuBisCO had come from over the preceding week. National Oceanic and Atmospheric Administration (NOAA) Air Resources Laboratory HYSPLIT back trajectory ensemble models92,93 were run using global data assimilation system (GDAS) meteorological data with a starting point at 30.62°N, 96.34°S (College Station, Texas, United States). Each model run started from the end of the 48 hr aerosol sampling period.
Statistical methods
All statistical methods were performed using JMP Pro 16 statistical software (JMP Statistical Discovery, LLC). Ice nucleation samples contain at least 5 replicates with the pooled mean and standard deviations reported (n ≤ 147). As these data did not meet the requirements of a parametric analysis of variance (ANOVA), a non-parametric one-way analysis ANOVA was performed to test the hypothesis that there were significant differences between median ice nucleation temperature using a two-tailed Kruskal–Wallis test on ranks. Pairwise post-hoc comparisons were made with the Wilcoxon test (p < 0.05).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data can be found in the Texas Data Repository (https://dataverse.tdl.org/): https://doi.org/10.18738/T8/VZAEK3.
References
Boucher, O. et al. Clouds and aerosols. In: Climate Change 2013 The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (eds. Stocker, T. F. et al.) 571-657 (Cambridge University Press, 2013).
Kanji, Z. A. et al. Overview of ice nucleating articles. Meteorol. Monogr. 58, 1.1–1.33 (2017).
Murray, B. J., O’Sullivan, D., Atkinson, J. D. & Webb, M. E. Ice nucleation by particles immersed in supercooled cloud droplets. Chem. Soc. Rev. 41, 6519–6554 (2012).
Brooks, S. D. & Thornton, D. C. O. Marine aerosols and clouds. Ann. Rev. Mar. Sci. 10, 289–313 (2018).
DeMott, P. J. et al. Predicting global atmospheric ice nuclei distributions and their impacts on climate. Proc. Natl. Acad. Sci. USA 107, 11217–11222 (2010).
Patade, S. et al. Empirical formulation for multiple groups of primary biological ice nucleating particles from field observations over Amazonia. J. Atmos. Sci. 78, 2195–2220 (2021).
Atkinson, J. D. et al. The importance of feldspar for ice nucleation by mineral dust in mixed-phase clouds. Nature 498, 355–358 (2013).
DeMott, P. J. et al. Sea spray aerosol as a unique source of ice nucleating particles. Proc. Natl. Acad. Sci. USA 113, 5797–5803 (2016).
Hoose, C. & Möhler, O. Heterogeneous ice nucleation on atmospheric aerosols: A review of results from laboratory experiments. Atmos. Chem. Phys. 12, 9817–9854 (2012).
Brooks, S. D., Suter, K. & Olivarez, L. Effects of chemical aging on the ice nucleation activity of soot and polycyclic aromatic hydrocarbon aerosols. J. Phys. Chem. A 118, 10036–10047 (2014).
Kanji, Z. A., Welti, A., Corbin, J. C. & Mensah, A. A. Black carbon particles do not matter for immersion mode ice nucleation. Geophys. Res. Lett. 47, e2019GL086764 (2020).
DeMott, P. J. An exploratory study of ice nucleation by soot aerosols. J. Appl. Meteorol. Climatol. 29, 1072–1079 (1990).
DeMott, P. J., Chen, Y., Kreidenweis, S. M., Rogers, D. C. & Sherman, D. E. Ice formation by black carbon particles. Geophys. Res. Lett. 26, 2429–2432 (1999).
Diehl, K., Matthias-Maser, S., Jaenicke, R. & Mitra, S. K. The ice nucleating ability of pollen: Part II. Laboratory studies in immersion and contact freezing modes. Atmos. Res. 61, 125–133 (2002).
Pummer, B. G., Bauer, H., Bernardi, J., Bleicher, S. & Grothe, H. Suspendable macromolecules are responsible for ice nucleation activity of birch and conifer pollen. Atmos. Chem. Phys. 12, 2541–2550 (2012).
Burkart, J., Gratzl, J., Seifried, T. M., Bieber, P. & Grothe, H. Isolation of subpollen particles (SPPs) of birch: SPPs are potential carriers of ice nucleating macromolecules. Biogeosciences 18, 5751–5765 (2021).
Gute, E. & Abbatt, J. P. D. Oxidative processing lowers the ice nucleation activity of birch and alder pollen. Geophys. Res. Lett. 45, 1647–1653 (2018).
Tesson, S. V. M. & Šantl-Temkiv, T. Ice nucleation activity and aeolian dispersal success in airborne and aquatic microalgae. Front. Microbiol. 9, 2681 (2018).
Wilbourn, E. K. et al. Ice nucleation by marine aerosols over the North Atlantic Ocean in late spring. J. Geophys. Res-Atmos 125, e2019JD030913 (2020).
Creamean, J. M. et al. Evaluating the potential for Haloarchaea to serve as ice nucleating particles. Biogeosciences 18, 3751–3762 (2021).
Maki, L. R., Galyan, E. L., Chang-Chien, M. M. & Caldwell, D. R. Ice nucleation induced by Pseudomonas syringae. Appl. Microbiol. 28, 456–459 (1974).
Failor, K. C., Schmale, D. G., Vinatzer, B. A. & Monteil, C. L. Ice nucleation active bacteria in precipitation are genetically diverse and nucleate ice by employing different mechanisms. ISME J 11, 2740–2753 (2017).
Fröhlich-Nowoisky, J. et al. Ice nucleation activity in the widespread soil fungus Mortierella alpina. Biogeosciences 12, 1057–1071 (2015).
Pouleur, S., Richard, C., Martin, J.-G. & Antoun, H. Ice nucleation activity in Fusarium acuminatum and Fusarium avenaceum. Appl. Environ. Microbiol. 58, 2960–2964 (1992).
Hill, T. C. J. et al. Sources of organic ice nucleating particles in soils. Atmos. Chem. Phys. 16, 7195–7211 (2016).
Adams, M. P. et al. Ice nucleation by viruses and their potential for cloud glaciation. Biogeosciences 18, 4431–4444 (2021).
Després, V. R. et al. Primary biological aerosol particles in the atmosphere: A review. Tellus B Chem. Phys. Meteorol. 64, 15598 (2012).
Levin, Z. & Yankofsky, S. A. Contact versus immersion freezing of freely suspended droplets by bacterial ice nuclei. J. Appl. Meteorol. Climatol. 22, 1964–1966 (1983).
Yankofsky, S. A., Nadler, T. & Kaplan, H. The presence of complete but masked freezing nuclei in various artificially constructed ice nucleation-active Proteobacteria. Curr. Microbiol. 34, 318–325 (1997).
Welti, A., Lüönd, F., Stetzer, O. & Lohmann, U. Influence of particle size on the ice nucleating ability of mineral dusts. Atmos. Chem. Phys. 9, 6705–6715 (2009).
Lienhard, D. M. et al. Viscous organic aerosol particles in the upper troposphere: diffusivity-controlled water uptake and ice nucleation? Atmos. Chem. Phys. 15, 13599–13613 (2015).
Collier, K. N. & Brooks, S. D. Role of organic hydrocarbons in atmospheric ice formation via contact freezing. J. Phys. Chem. A 120, 10169–10180 (2016).
Roeters, S. J. et al. Ice-nucleating proteins are activated by low temperatures to control the structure of interfacial water. Nat. Commun. 12, 1183 (2021).
Wang, X. et al. Microbial control of sea spray aerosol composition: a tale of two blooms. ACS Cent. Sci. 1, 124–131 (2015).
Wilson, T. W. et al. A marine biogenic source of atmospheric ice-nucleating particles. Nature 525, 234–238 (2015).
Ickes, L. et al. The ice-nucleating activity of Arctic sea surface microlayer samples and marine algal cultures. Atmos. Chem. Phys. 20, 11089–11117 (2020).
Alpert, P. A., Aller, J. Y. & Knopf, D. A. Ice nucleation from aqueous NaCl droplets with and without marine diatoms. Atmos. Chem. Phys. 11, 5539–5555 (2011).
Knopf, D. A., Alpert, P. A., Wang, B. & Aller, J. Y. Stimulation of ice nucleation by marine diatoms. Nat. Geosci. 4, 88–90 (2011).
Armbrust, E. V. et al. The genome of the diatom Thalassiosira Pseudonana: Ecology, evolution, and metabolism. Science 306, 79–86 (2004).
Tobo, Y. et al. Biological aerosol particles as a key determinant of ice nuclei populations in a forest ecosystem. J. Geophys. Res-Atmos. 118, 10–100 (2013).
Hiranuma, N. et al. Ice nucleation by cellulose and its potential contribution to ice formation in clouds. Nat. Geosci. 8, 273–277 (2015).
Hiranuma, N. et al. A comprehensive characterization of ice nucleation by three different types of cellulose particles immersed in water. Atmos. Chem. Phys. 19, 4823–4849 (2019).
Bogler, S. & Borduas-Dedekind, N. Lignin’s ability to nucleate ice via immersion freezing and its stability towards physicochemical treatments and atmospheric processing. Atmos. Chem. Phys. 20, 14509–14522 (2020).
Wolf, M. J. et al. Investigating the heterogeneous ice nucleation of sea spray aerosols using Prochlorococcus as a model source of marine organic matter. Environ. Sci. Technol. 53, 1139–1149 (2019).
Wex, H. et al. Intercomparing different devices for the investigation of ice nucleating particles using Snomax® as test substance. Atmos. Chem. Phys. 15, 1463–1485 (2015).
Ellis, R. J. The most abundant protein in the world. Trends Biochem. Sci. 4, 241–244 (1979).
Raven, J. A. Rubisco: Still the most abundant protein of Earth? New Phytol. 198, 1–3 (2013).
Bar-On, Y. M. & Milo, R. The global mass and average rate of rubisco. Proc. Natl. Acad. Sci. USA 116, 4738–4743 (2019).
Valegård, K., Hasse, D., Andersson, I. & Gunn, L. H. Structure of Rubisco from Arabidopsis thaliana in complex with 2-carboxyarabinitol-1,5-bisphosphate. Acta Crystallogr. Sect. D-Struct. Biol. D 74, 1–9 (2018).
Losh, J. L., Young, J. N. & Morel, F. M. M. Rubisco is a small fraction of total protein in marine phytoplankton. New Phytol. 198, 52–58 (2013).
Fornea, A. P., Brooks, S. D., Dooley, J. B. & Saha, A. Heterogeneous freezing of ice on atmospheric aerosols containing ash, soot, and soil. J. Geophys. Res. Atmos. 114, D13201 (2009).
Service, R. F. Protein structures for all. Science 374, 1426–1427 (2021).
Forman, H. J., Zhang, H. & Rinna, A. Glutathione: Overview of its protective roles, measurement, and biosynthesis. Mol. Aspects. Med. 30, 1–12 (2009).
Meister, A. Gluthathione metabolism and its selective modification. J. Biol. Chem. 263, 17205–17208 (1988).
Cascajo-Castresana, M., David, R. O., Iriarte-Alonso, M. A., Bittner, A. M. & Marcolli, C. Protein aggregates nucleate ice: The example of apoferritin. Atmos. Chem. Phys. 20, 3291–3315 (2020).
Rastelli, E. et al. Transfer of labile organic matter and microbes from the ocean surface to the marine aerosol: An experimental approach. Sci. Rep. 7, 1–10 (2017).
Cáliz, J., Triadó-Margarit, X., Camarero, L. & Casamayor, E. O. A long-term survey unveils strong seasonal patterns in the airborne microbiome coupled to general and regional atmospheric circulations. Proc. Natl. Acad. Sci. USA 115, 12229–12234 (2018).
Alsante, A. N., Thornton, D. C. O. & Brooks, S. D. Ocean Aerobiology. Front. Microbiol. 12, 764178 (2021).
Lang-Yona, N. et al. Terrestrial and marine influence on atmospheric bacterial diversity over the north Atlantic and Pacific Oceans. Commun. Earth Environ. 3, 1–10 (2022).
Chen, J. et al. Atmospheric humic-like substances (HULIS) act as ice active entities. Geophys. Res. Lett. 48, e2021GL092443 (2021).
Gromiha, M. M. Protein Bioinformatics: From Sequence to Function (Elsevier Science, 2010).
Garnham, C. P., Campbell, R. L., Walker, V. K. & Davies, P. L. Novel dimeric β-helical model of an ice nucleation protein with bridged active sites. BMC Struct. Biol. 11, 1–12 (2011).
Hudait, A., Odendahl, N., Qiu, Y., Paesani, F. & Molinero, V. Ice-nucleating and antifreeze proteins recognize ice through a diversity of anchored clathrate and ice-like motifs. J. Am. Chem. Soc. 140, 4905–4912 (2018).
Eickhoff, L. et al. Contrasting behavior of antifreeze proteins: Ice growth inhibitors and ice nucleation promoters. J. Phys. Chem. Lett. 10, 966–972 (2019).
Kim, T. D., Ryu, H. J., Cho, H. I., Yang, C. H. & Kim, J. Thermal behavior of proteins: heat-resistant proteins and their heat-induced secondary structural changes. Biochem. 39, 14839–14846 (2000).
Salvucci, M. E., Osteryoung, K. W., Crafts-Brandner, S. J. & Vierling, E. Exceptional sensitivity of Rubisco activase to thermal denaturation in vitro and in vivo. Plant Physiol. 127, 1053–1064 (2001).
Subramaniam, V., Bergenhem, N. C. H., Gafni, A. & Steel, D. G. Phosphorescence reveals a continued slow annealing of the protein core following Rreactivation of Escherichia coli alkaline phosphatase. Biochem. 34, 1133–1136 (1995).
Wright, R. C. & Tramer, J. Reactivation of milk phosphatase following heat treatment. I. J. Dairy Res. 20, 177–188 (1953).
Wright, R. C. & Tramer, J. Reactivation of milk phosphatase following heat treatment. II. J. Dairy Res. 20, 258–273 (1953).
Steinke, I. et al. A numerical framework for simulating the atmospheric variability of supermicron marine biogenic ice nucleating particles. Atmos. Chem. Phys. 22, 847–859 (2022).
Daily, M. I., Tarn, M. D., Whale, T. F. & Murray, B. J. An evaluation of the heat test for the ice-nucleating ability of minerals and biological material. Atmos. Meas. Tech. 15, 2635–2665 (2022).
Bischof, J. C. & He, X. Thermal stability of proteins. Ann. Ny. Acad. Sci. 1066, 12–33 (2006).
Petters, M. D. & Wright, T. P. Revisiting ice nucleation from precipitation samples. Geophys. Res. Lett. 42, 8758–8766 (2015).
Orellana, M. V. & Hansell, D. A. Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO): A long-lived protein in the deep ocean. Limnol. Oceanogr. 57, 826–834 (2012).
Møller, E. F. Production of dissolved organic carbon by sloppy feeding in the copepods Acartia tonsa, Centropages typicus, and Temora longicornis. Limnol. Oceanogr. 52, 79–84 (2007).
Møller, E. F., Thor, P. & Nielsen, T. G. Production of DOC by Calanus finmarchicus, C. glacialis and C. hyperboreus through sloppy feeding and leakage from fecal pellets. Mar. Ecol. Prog. Ser. 262, 185–191 (2003).
Bratbak, G., Egge, J. K. & Heldal, M. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar. Ecol. Prog. Ser. 93, 39–48 (1993).
Mojica, K. D. A., Huisman, J., Wilhelm, S. W. & Brussaard, C. P. D. Latitudinal variation in virus-induced mortality of phytoplankton across the North Atlantic Ocean. ISME J. 10, 500–513 (2016).
Russell, L. M., Hawkins, L. N., Frossard, A. A., Quinn, P. K. & Bates, T. S. Carbohydrate-like composition of submicron atmospheric particles and their production from ocean bubble bursting. Proc. Natl. Acad. Sci. USA 107, 6652–6657 (2010).
O’Dowd, C. D. et al. Biogenically driven organic contribution to marine aerosol. Nature 431, 676–680 (2004).
Quinn, P. K., Collins, D. B., Grassian, V. H., Prather, K. A. & Bates, T. S. Chemistry and related properties of freshly emitted sea spray aerosol. Chem. Rev. 115, 4383–4399 (2015).
Tesson, S. V. M., Skjøth, C. A., Šantl-Temkiv, T. & Löndahl, J. Airborne Microalgae: Insights, opportunities, and challenges. Appl. Environ. Microbiol. 82, 1978–1991 (2016).
Sherwood, A. R., Wade, R. M. & Conklin, K. Y. Seasonality of tropical airborne algae: a 16-month study based on high-throughput sequencing in the Hawaiian Islands. Grana 59, 354–365 (2020).
Jassey, V. E. J. et al. Contribution of soil algae to the global carbon cycle. New Phytol. 234, 64–76 (2022).
Bianco, A. et al. Effect of endogenous microbiota on the molecular composition of cloud water: a study by Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR MS). Sci. Rep. 9, 1–12 (2019).
Zelinka, M. D., Randall, D. A., Webb, M. J. & Klein, S. A. Clearing clouds of uncertainty. Nat. Clim. Chang. 7, 674–678 (2017).
Tabita, F. R., Satagopan, S., Hanson, T. E., Kreel, N. E. & Scott, S. S. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 59, 1515–1524 (2008).
Doyle, S. M. et al. Rapid formation of microbe-oil aggregates and changes in community composition in coastal surface water following exposure to oil and the dispersant corexit. Front. Microbiol. 9, 689 (2018).
Lee, M. D. et al. Marine Synechococcus isolates representing globally abundant genomic lineages demonstrate a unique evolutionary path of genome reduction without a decrease in GC content. Environ. Microbiol. 21, 1677–1686 (2019).
Yona, A. H., Alm, E. J. & Gore, J. Random sequences rapidly evolve into de novo promoters. Nat. Commun. 9, 1–10 (2018).
Vali, G., DeMott, P. J., Möhler, O. & Whale, T. F. Technical Note: a proposal for ice nucleation terminology. Atmos. Chem. Phys 15, 10263–10270 (2015).
Stein, A. F. et al. NOAA’s HYSPLIT atmospheric transport and dispersion modeling system. Bull. Am. Meteorol. Soc. 96, 2059–2077 (2015).
Rolph, G., Stein, A. & Stunder, B. Real-time environmental applications and display sYstem: READY. Environ. Model. Softw. 95, 210–228 (2017).
Acknowledgements
Financial support was provided by NSF Atmospheric Chemistry Program (Award # AGS-2128133) to D.C.O.T. and S.D.B.. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. We thank Shawn M. Doyle and Jason B. Sylvan of the Department of Oceanography, Texas A&M University, for their guidance on RNA sample preparation and the use of the plate reader for RuBisCO quantification. The authors gratefully acknowledge the NOAA Air Resources Laboratory (ARL) for the provision of the HYSPLIT transport and dispersion model and READY website (https://www.ready.noaa.gov) used in this publication.
Author information
Authors and Affiliations
Contributions
A.N.A. carried out sample preparation, measurements, and data analysis in consultation with D.C.O.T. and S.D.B.. A.N.A., D.C.O.T. and S.D.B. contributed to the writing of the manuscript. D.C.O.T. and S.D.B. designed the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Earth & Environment thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Yinon Rudich, Clare Davis, Heike Langenberg.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alsante, A.N., Thornton, D.C.O. & Brooks, S.D. Ice nucleation catalyzed by the photosynthesis enzyme RuBisCO and other abundant biomolecules. Commun Earth Environ 4, 51 (2023). https://doi.org/10.1038/s43247-023-00707-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43247-023-00707-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.