Abstract
The diversity of microbes and their transmission between ocean and atmosphere are poorly understood despite the implications for microbial global dispersion and biogeochemical processes. Here, we survey the genetic diversity of airborne and surface ocean bacterial communities sampled during springtime transects across the northwest Pacific and subtropical north Atlantic as part of the Tara Pacific Expedition. We find that microbial community composition is more variable in the atmosphere than in the surface ocean. Bacterial communities were more similar between the two surface oceans than between the ocean and the overlying atmosphere. Likewise, Pacific and Atlantic atmospheric microbial communities were more similar to each other than to those in the ocean beneath. Atmospheric community composition over the Atlantic was dominated by terrestrial and specifically, dust-associated bacteria, whereas over the Pacific there was a higher prevalence and differential abundance of marine bacteria. Our findings highlight regional differences in long-range microbial exchange and dispersal between land, ocean, and atmosphere.
Similar content being viewed by others
Introduction
Microbes are ubiquitous in oceanic and atmospheric environments. Within the ocean, they account for approximately 70% of the total marine biomass1, playing a crucial role in biogeochemical cycles (such as the carbon, nitrogen, and sulfur cycles)2. In the atmosphere, the microbial community comprises a major part of atmospheric bioaerosols3, but little is known about the factors that affect their diversity, abundance4, and role in the air5, as well as the microbial exchange between the two environments6.
Understanding the ocean's role as a source and sink of microorganisms and the transport of airborne bacteria can provide important insights into microbial spatial distribution, diversity, and the interplay between terrestrial communities and their transmission over oceanic regions. Overland, airborne bacteria are emitted from a wide range of sources, from anthropogenic to natural ecosystems. Over the oceans, bacteria can be locally emitted as sea spray aerosols, generated at the ocean’s surface by wind-driven processes, or transferred into the marine atmosphere through long-range transport from terrestrial sources4. Due to their aerodynamic sizes, it has been hypothesized that all airborne bacteria can disperse globally and may proliferate in any habitat with suitable environmental conditions7,8. Thus, it can be expected that the airborne bacterial community aloft in a given marine environment would exhibit a large fraction of the local marine microbiome.
Indeed, biodiversity studies indicated a link between microbial community composition and geographic locations8,9,10. Nevertheless, the geo-distribution of microorganisms in the interface between the ocean and the atmosphere is still mostly underexplored, as the type, amount, and efficiency of particle emissions from the ocean or atmosphere deposition hold large uncertainties. Exploring such distributions and microbial fluxes can improve our understanding of the metabolic capabilities introduced into the ocean and the enrichment of local marine diversity11,12. They may also affect biogeochemical processes upon deposition of bacteria into the ocean or their release into the atmosphere.
In this study, we present the spatially resolved composition of bacterial communities in the atmospheric marine boundary layer (AMBL) and the ocean surface along two basin-scale oceanic transects: The North Atlantic Ocean and the Western Pacific Ocean, covering approximately 15,000 km. The similar seasons during which sampling occurred, both located in the northern hemisphere (except for a few sampling points in the Pacific Ocean, south of the Equator), allow us to focus on non-seasonal changes in the microbial communities across these transects. We first explore microbial community compositions in the atmospheric and oceanic environments, revealing larger similarities between different oceans, in contrast to their overlaying atmospheres. By focusing on specific genera, we track terrestrial-associated bacteria in the airborne community and show distinct patterns of marine bacterial emission into the atmosphere. Finally, we suggest that theories and the attempt for estimating microbial distribution between environments should include constraints between the hydrosphere and atmosphere.
Results and discussion
Regional distribution of airborne and surface water bacterial phyla in the Pacific and Atlantic oceans
The two open ocean sailing transects examined in this study included the western Pacific path, sampled in May 2017 from Keelung, Taiwan, towards Fiji (Fig. 1a and Supplementary Data 1), and the Atlantic crossing, sampled in June 2016 from Lorient, France, to Miami, USA (Fig. 1b and Supplementary Data 1). In the water, we found a higher homogeneity in phyla distribution within each transect (significantly lower Euclidean distances between centered log-ratio (CLR)-converted phyla counts (betadispar): Atlantic: 0.3212 compared to 0.4229 in the air, ANOVA (with Tukey’s post hoc), p < 0.0001; Pacific: 0.1865 compared to 0.3891 in the air, ANOVA (Tukey’s post hoc), p < 0.0001; Tables S1–S4), with Proteobacteria dominating both oceans (58 ± 3% in the Pacific and 66 ± 4% in the Atlantic; Fig. 1c, d, respectively). Cyanobacteria (29 ± 2% and 9 ± 4%) and Bacteroidetes (9 ± 1% and 17 ± 2%) were the next two most abundant phyla. When compared to other marine microbiome studies, the phyla distribution of the near-surface water environment was similar2,13. For example, the Cyanobacteria to Proteobacteria ratios in our study are 0.49 ± 0.06 and 0.14 ± 0.07 in the Pacific and Atlantic surface water, respectively. Similarly-calculated ratios characterized in these oceanic regions were 0.43 and 0.16, for the Pacific13 and Atlantic2 regions, respectively.
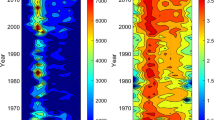
Air-sampling locations and back trajectories, with height above mean sea levels (MSL) indicated with color scale for the Pacific (a) and Atlantic (b) atmospheric samples. Phyla differential abundance (centered log-ratio; CLR- converted abundance) in the water (c, d) and the air (e, f) for taxa observed in >5% of the samples in the Atlantic and Pacific transects, respectively.
In the AMBL, Proteobacteria was also the most dominant airborne bacterial phylum in both the Pacific and Atlantic oceans, with 69 ± 12% and 64 ± 8% average percentile abundance, respectively (Fig. 1e, f). However, we found a more heterogeneous distribution of bacterial phyla than in ocean surface water samples (Fig. S1), even when considering air masses spending at least 120 h over an oceanic path prior to sampling (Fig. 1a, b, colored lines). Other abundant phyla in the Pacific AMBL included Cyanobacteria (11 ± 9%), Bacteroidetes (8 ± 3%), Firmicutes (8 ± 3%), Actinobacteria (4 ± 3%), and Planctomycetes (2 ± 1%). In the Atlantic AMBL, the abundant phyla included Actinobacteria (11 ± 5%), Firmicutes (10 ± 6%), and Bacteroidetes (6 ± 3%; Fig. 1f), while Cyanobacteria was observed with an average abundance of less than 0.5%. In general, while Proteobacterial abundance is high in both air and water, there is a distinct difference in Bacteroidetes (higher in oceanic samples, Two-sample t-test, performed on CLR values, equal variance (F = 1.174, p = 0.341), t = 14.889; p = 1.91 × 10−22).
Firmicutes were predominantly observed in the air samples (8 ± 3% and 10 ± 6% in the Pacific and Atlantic AMBL, respectively), with low (<1% in average) to non-significant abundance in the water samples (unequal variance t-test on CLR values (F = 5171.823, p = 1.2 × 10−83), t = 25.716; p = 2.1 × 10−36). Actinobacteria abundance was also significantly higher in the Atlantic AMBL compared to the water (unequal variance t-test on CLR values (F = 68.383, p = 4.6 × 10−35), t = 17.117, p = 1.2 × 10−26). Airborne Actinobacteria and Firmicutes have been previously connected to desert dust samples in the Eastern Mediterranean14,15,16, and detected in different studies of airborne bacteria17,18,19. In addition, Firmicutes are usually more abundant in soils than in marine surface water2,20, where specifically the Bacillus genus was detected20. Sul et al. found marine Firmicutes in low relative abundance (<6% on average) across latitudes with little latitudinal dependence21. Therefore, the airborne Firmicutes most likely represent a terrestrial source.
Members of the airborne-detected Fusobacteria, Deinococcus-Thermos, and Acidobacteria phyla (Fig. 1e, f) exhibit high physiological diversity in cell shapes and sizes20,22. Their presence in remote locations, such as Antarctica23, airborne dust14, and precipitation over the alpine24, together with their detection in the current study above the western Pacific Ocean, after 120 h transport above the ocean, suggests that they are ubiquitous in the atmospheric environment.
The local primary production impact on the AMBL was estimated by calculating the ratio of known autotrophic to heterotrophic bacterial amplicon sequence variant (ASV; as listed in Table S5)25 in the atmospheric and oceanic samples (Fig. S2). The ratios in the Pacific AMBL were more than an order of magnitude higher than those measured in the Atlantic AMBL (mean values ± SE: 0.186 ± 0.029 and 0.005 ± 0.002, respectively, Wilcoxon rank test, p = 7.44 × 10−06; Fig. S2a, b). Additionally, the average ratio in the Pacific surface water is approximately four times higher than in the Atlantic (0.406 ± 0.010, compared to 0.104 ± 0.010, respectively; Wilcoxon rank test, p = 2.98 × 10−08). Similarly, a significantly higher relative abundance of cyanobacterial 16S rRNA gene to total bacterial 16S rRNA gene was observed in the Pacific compared to the Atlantic air samples (mean values ± SE: 0.171 ± 0.016 and 0.001 ± 0.000, respectively, Wilcoxon rank test, p = 1.863 × 10−09; Fig. S2c, d and Table S6) based on qPCR analysis (Supplementary Note 8). The observed difference in oceanic cyanobacterial abundance is consistent with results reported by Flombaum et al., showing a higher abundance of marine Synechococcales in the Pacific compared to the Atlantic26.
Similarities and differences in the atmospheric and oceanic microbiomes
The marine and atmospheric microbiomes were further analyzed at higher taxonomic resolution. We found the airborne bacterial diversity to be significantly higher compared to the water samples (Fig. 2a; DivNet for Shannon diversity, Wilcoxon rank test, p = 2.98 × 10−08 for the Atlantic, and p = 2.587 × 10−05 for the Pacific), as well as the airborne bacterial composition distances (Fig. 2b; ANOSIM R = 0.7519, p = 0.01). In addition, both mean Euclidean distances (Fig. 2c; Kruskal–Wallis chi-squared = 1321.9, df = 1, p < 2.2 × 10−16; Wilcoxon rank test, p < 2.2 × 10−16 for both Atlantic and Pacific environments) show that for each environment, the surface water sample distances are significantly smaller compared to the AMBL samples, indicating a more homogeneous and stable community structure in the marine surface water.
Biome-based diversity (based on the Inverse Simpson index) of amplicon sequence variants (ASVs) detected in more than 5% of the samples (a) and clusters represented by nonmetric multidimensional scaling (NMDS) ordination, based on Bray-Curtis dissimilarity metrics (b), with 95% confidence ellipses. The distance in ASV composition is presented in a heatmap of the Bray-Curtis dissimilarity between all analyzed samples (Bray-Curtis index varies between 0, for an identical ASV composition, and 1, for most distant ASVs between samples) (c). A Venn diagram (VennDiagram 1.6.20) summarizes the number and the percentage (in brackets) out of the total number of bacterial taxa that were observed in the different biomes (d). Box plot center lines represent the median values, box limits are the 25 and 75% of the population, upper and lower quartiles represented in bars are 5 and 95% of the population; whiskers represent significance <0.05, points at (c) represent outliers.
The surface water environments shared 166 taxa between the Atlantic and Pacific oceans, and the AMBL biomes shared 341 taxa (comprising 28% of all ocean taxa vs. 25% of all airborne taxa; Fig. 2d). Only 78 taxa in the Atlantic and 134 in the Pacific (7% of all Atlantic taxa vs. 15% of all Pacific taxa) were shared between an ocean and its corresponding atmosphere. The ubiquity of shared species found only in the atmospheric samples of the Atlantic and Pacific oceans suggests a potentially higher pool of air-resident bacteria with efficient long-range transport in the atmosphere. In addition, different oceans are found to have a greater resemblance to one another than to their overlaying AMBL, and atmospheric samples from distinct locations (at least 13,000 km apart from each other) share more common taxa than the ocean beneath. This suggests that the proximity of the sampled biomes is second in significance to the type of sampled environment.
A phylogenetic tree based on the bacterial 16S amplicon sequences provides an overview of the bacterial community and genetic distances between them in the observed marine environment (Fig. S3). Among the shared groups in the Atlantic and Pacific atmospheric biomes, the main phyla occurrences included 44% Proteobacteria, 19% Actinobacteria, 19% Firmicutes, and 10% Bacteroidetes.
While the wind-driven surface water currents show connectivity between oceans with time scales of years, the atmospheric circulation time scales are in the range of days to weeks27,28. Therefore, the high diversity and variations between samples of airborne bacteria in both the Atlantic and Pacific is most likely a consequence of a short turnover time of the air mass, leading to continuously changing and dynamic community composition in the AMBL.
Spatial distributions of bacteria across the Pacific and Atlantic environments
To better understand possible nonrandom exchanges between the ocean and the atmosphere, we explored the association between bacterial taxa and geographic locations (Atlantic vs. Pacific), as well as different environments (air vs. water). The differential (CLR-converted) abundance of the marine taxa detected in the pacific AMBL samples was 19.5%, while only 2.6% in the Atlantic AMBL (Fig. 3a). The highest bacterial prevalence in the AMBL samples was observed for Pseudomonas (with 88 and 64% prevalence in air and water, respectively), continuously detected in the two environments. This taxon was significantly classified as associated with the air environment (Taxon 09 in Fig. 3a and Supplementary Data 4, MaAsLin2 Coeff. = 2.570, q < 0.0001). The contribution of bacteria to the formation of water precipitation is of high interest, and studies revealed bacterial proteins e.g., Pseudomonas sp., can promote droplet freezing29 and even detected bacterial activity in clouds19. Paracoccus, a Proteobacterium detected only in the AMBL, was found in 84% of air samples, with a significant association with the air environment (Taxon 22 in Fig. 3a and Supplementary Data 4, MaAsLin2 Coeff. = 2.457, q < 0.0001). Paracoccus strains have been isolated from different environments including soil30, marine sediments31, sewage32, and have been detected in other atmospheric bacterial studies33,34.
The 100 Bacterial taxa, with the highest score and significance of <0.05 for association with a specific environment (Atlantic or Pacific; air or water; tested using MaAsLin2 for multivariable association discovery in population-scale studies) are clustered based on phylogenetic similarity and scaled in a heatmap based on centered log-ratio (CLR)-converted abundance (a). Highlighted taxa in the text are indicated with an asterisk and specified in Supplementary Data 5. The MaAsLin2 score per phylum are presented with association to water and air (b), and Atlantic and Pacific (c). Error bars represent the standard error of the MaAsLin2 coefficient.
The most observed Firmicutes genus that appeared exclusively in the air samples was Bacillus, found in 81% of air samples, with significant association with the Pacific air environment (Taxon 23 in Fig. 3a and Supplementary Data 4, MaAsLin2 Coeff. = 2.717, and 1.261 for air and the pacific associations, respectively, q < 0.0001 in both cases). This genus is known to contain endospores that can remain dormant for years. Bacillus is a common bacterium found in transported desert dust14,35, and the deep marine environment36. Some Bacillus species are known for their unique metabolites and antagonistic activity against pathogens37. The rare abundance of marine bacilli in the water may result from the preferential growth environment of the deep sea, coral, and sediments36, and their copiotrophic property (i.e., flourishing in environments with high nutrient availability)38. Notably, additional spore-forming Firmicutes, e.g., Tumebacillus39, were detected in the Atlantic AMBL (Taxon 76 in Fig. 3a and Supplementary Data 5). Endospores can survive harsh and dry conditions and thus might be transported through the air at higher survival rates than others.
The underrepresentation of Firmicutes in the water samples was further explored in a focused phylogenetic analysis targeting only Firmicutes ASVs (Fig. S5a). The few detected water ASVs differed phylogenetically from the atmospheric ASVs. Additionally, in a supportive environmental ontology (ENVO) analysis40 on airborne Firmicutes ASVs in the Atlantic and Pacific (Fig. S5b, c and Supplementary Note 7), a higher relative contribution of soil-borne ENVO annotations was observed in both environments.
Since prokaryote concentrations in the atmosphere are orders of magnitude smaller compared to the ocean (~103–104 m−3 in the atmosphere4,41 vs. ~1011–1015 m−3 in the surface waters20,41), sedimentation of terrestrial-originated bacteria to the ocean are not expected to induce a significant change in the microbial diversity in the water, unless profound proliferation takes place. Thus, we conjecture that the airborne Firmicutes most probably originate from terrestrial long-range transport, but the extent to which their sedimentation and proliferation occur in the ocean is yet to be determined.
We have detected bacterial genera that are known to include human-associated microbes (i.e., Micrococcus, Actinomyces; Fig. 3a and Supplementary Data 5). Although not detected in the blank filters, we cannot exclude the possibility that those taxa may originate from the human activity onboard Tara. Nevertheless, the prevalence of these genera is low, ranging on average between ~0.1–~2.7% of the ASVs per filter.
The most prevailing Actinobacteria was Actinomarina, a marine bacterium, appearing in all oceanic samples and in the Pacific air samples (Taxon 05, in Fig. 3a and Supplementary Data 5). Its presence suggests that the surface water contributes to the overlying atmosphere, yet to a limited extent. The highest occurring Bacteroidetes genera found in the atmospheric samples were Chryseobacterium and Sediminibacterium (Taxa 82, and 45 in Fig. 3a, found in 43, and 40% of the air samples, respectively; Supplementary Data 4, 5). Sediminibacterium (MaAsLin2 Coeff. = 2.022 and 1.686 for air and the pacific associations, respectively, q < 0.0001 in both cases) was previously found to contribute to the coral microbiome42 and detected in air samples over the Great Barrier Reef17 and the Mediterranean Sea43. In our study, it was detected in 40% of the air samples, and in the water samples of the Pacific Ocean solely, with low relative abundance (<0.01%) and spatial prevalence.
We continuously detected water-borne species, known from the literature, in the air samples, with higher relative abundance in the Pacific AMBL. One such genus is the Cyanobacteria Prochlorococcus (Taxon 01 in Fig. 3a and Supplementary Data 5) considered a key and most abundant autotroph44, found mainly in oligotrophic oceans45, with 100% prevalence in the ocean compared to 54% in the air, from which 75% were in the Pacific AMBL (Wilcoxon rank test, p = 3.03 × 10−06).
Terrestrial-associated airborne bacteria
Key species found in our analysis to be significantly dominant in the Atlantic air (e.g., Paracoccus, Methylobacterium, Mesorhizobium, etc.; Fig. 3a), are known as terrestrial-46,47,48 and specifically, dust-35,49 associated bacteria. The ENVO annotation (Fig. S6) retrieved from genomic databases corroborates these findings and emphasizes a significantly higher terrestrial-annotated bacteria predicted to be a partial source for the Atlantic AMBL (Wilcoxon rank test, p = 1.49 × 10−08). Additionally, we detected significantly higher DNA biomass in the Atlantic air samples compared to the Pacific (average of 639.6 ± 468.2 and 128.4 ± 54.4 pg m−3, respectively; Two-sample t-test, unequal variance (F = 74.097, p < 0.001), t = 4.7288, p = 0.0002), implying a higher concentration of microbial cells per air volume in this region.
A previous study by Mayol et al.41 determined that overall, 25% of the airborne bacteria over the ocean originated from the marine environment, and 42% originated from terrestrial sources based on parameterizations of sea spray and deposition flux calculations. Flores et al.50,51 have found higher concentrations of larger particles related to the deposition of mineral dust in aerosols sampled in the Atlantic compared to the Pacific transect. Together with other studies reporting on massive dust quantities crossing over the Atlantic Ocean52,53, they support our findings indicating a vast contribution of terrestrial-borne and specifically, dust-associated bacteria into the Atlantic Ocean.
Selectivity in the emission of marine bacteria
While the increased fraction of terrestrial-associated bacteria in the Atlantic AMBL could partially explain the reduction in the relative abundance of the local marine bacteria, marine-associated taxa were absent from a significantly high fraction of the sampled Atlantic AMBL, suggesting other factors may also play a role in the observed difference between the two AMBLs. A clear case of such difference is seen for the Pelagibacterales (SAR-11 clade; (Taxon 03, 06, 08, and others in Fig. 3a and Supplementary Data 5), representing approximately one-third of the oceanic surface water microbial community54, and highly abundant in both oceans’ samples. Their prevalence ratio between atmospheric and oceanic samples is significantly lower than Prochlorococcus (Wilcoxon rank test for the Atlantic (p = 0.006), and Pacific (p = 4.847 × 10−06); Supplementary Data 4). However, while the prevalence is minimal in the Atlantic, the Pacific AMBL shows a 79% prevalence of SAR-11 ASVs in these samples, significantly higher than the Atlantic (Wilcoxon signed-rank test, p = 3.182 × 10−05; Supplementary Data 4). A reduced aerosolized fraction of SAR-11 compared to the seawater was also observed in the Arctic Sea by Fahlgren et al.55. Part of the difference in abundance between the Atlantic and Pacific AMBLs could be related to properties of the sea-surface microlayer, including thickness, concentration, and chemical composition, which was shown to differ according to changes in heat exchanges, microbial composition, oceanic waves, pollution, and dust storms56,57. However, to fundamentally characterize the causing factors for the reduced detection of marine bacteria in the Atlantic AMBL, further investigation is required.
Possible boundaries between air and water
Environmental associations between air and water (Fig. 3b) were found to be more profound than those between the Atlantic and Pacific environments (Fig. 3c). This strengthens our findings that the type of the environment (air vs. water) is higher in contribution compared to the geographic location (Atlantic vs. Pacific) in influencing the microbial composition. A notable difference in the abundance and prevalence of marine bacteria in the Pacific AMBL compared to the Atlantic AMBL is also seen for different phyla, including Bacteroidetes, Verrucomicrobia, and Planctomycetes (Fig. 3c). Supportive evidence derived from ENVO analysis of the Atlantic and Pacific AMBLs similarly indicates a higher fraction of marine-annotated bacteria in the Pacific (Fig. S6a, b; 47 ± 20% compared to 12 ± 5% in the Atlantic), while the Atlantic aerobiome was dominantly predicted to be from a terrestrial origin (Fig. S6c, d; 59 ± 16% compared to 35 ± 18% in the Pacific).
It seems that boundaries might be drawn between the atmosphere and hydrosphere, allowing a nonrandom distribution of species between them. One case is the underrepresentation of marine bacteria in the Atlantic air, and others are airborne taxa (e.g., Firmicutes species) not detected in the water samples. Zhou & Ning (2017)10 have reviewed stochastic vs. deterministic mechanisms controlling microbial ecology and environmental processes that impact the balance between the two mechanisms. However, the effect of environmental interphases, such as the sea-surface microlayer on microbial biogeography and their impact on selective transmission from the ocean to the atmosphere, is still underexplored. We, therefore, propose that the chemical and physical properties of the sea-surface microlayers may determine in part the extent of selective transport of different bacteria from the ocean to the atmosphere.
Conclusions
This study illustrates microbial landscape in the interface between the ocean and the atmospheric boundary layer and their spatial diversity and suggests mechanisms of dispersal at both local and global scales (Fig. 4). We observed a significant difference in microbial diversity between the atmospheric and oceanic microbiomes, across thousands of kilometers in the north Atlantic and western Pacific oceans. A highly variable airborne microbial composition was detected (Fig. 4a), compared to a relatively more stable, and homogeneous composition of the surface-water microbiome, even across latitudes and different oceans (Fig. 4b). The contrast in microbial composition fluctuations may be attributed to the orders of magnitude differences in the characteristic advection and mixing scales in the two media. The oceanic microbial composition can be influenced by additional short-scale scenarios e.g., microalgal blooms, extreme weather events, anthropogenic pollution, as well as seasonal differences, but was not captured in this survey.
Higher distances between microbial composition in air samples is probably induced by residence time rather than by physical mixing (a, b). The microbial compositions of similar environments (e.g., Atlantic and Pacific Oceans) are more similar than geographical relations (e.g., Pacific atmosphere and surface water; c). The marine bacterial aerosolization seems to be selective (d) and differs in efficiency for different oceans (e). The atmospheric contribution to the marine ecosystem was not detected in the framework of this study (f), but it cannot be excluded that differences in atmospheric microbial composition could impact deposition and surface water composition and function. A terrestrial signature was detected in a remote oceanic environment (g), with a significant contribution of terrestrial- and dust-associated bacteria in the Atlantic atmospheric marine boundary layer compared to the Pacific (h).
The Atlantic and Pacific water samples showed greater resemblance to one another than to the atmosphere atop, and thousands of kilometers distant atmospheric samples shared more common taxa than the ocean beneath (Fig. 4c). This suggests that the proximity of the sampled biomes might be less influential compared to the unique ecosystem (i.e., water vs. air).
Differences in the differential abundance of marine bacteria were observed in the Pacific compared to the Atlantic AMBL, with a significant reduction in their prevalence in the Atlantic (Fig. 4d). This observation is probably not linked with a dilution effect of additional terrestrial particulates in the Atlantic AMBL, as these taxa were consistently low in appearance in the Atlantic air and presented high sequencing coverage (Fig. S13). Differences in the properties of these environments and of their sea-surface microlayers could impact the aerosolization efficiency of local marine bacteria and explain the differences in their atmospheric abundance, but this hypothesis requires further validation. Although reduced in the Atlantic air, the airborne autotrophic cyanobacteria maintained a similar ratio vs. heterotrophs as in the surface waters in both environments, while other species (e.g., SAR-11) were significantly reduced in the Atlantic AMBL, although highly abundant in the surface waters (Fig. 4e). This difference in the air to water ratio might suggest a selectivity in the emission of specific bacterial species. Therefore, theories and future estimates of microbial distribution between environments should include constraints between the ocean surface and its overlaying atmosphere.
While bacterial taxa associated with the terrestrial environment (e.g., Firmicutes) were found to continuously appear in the AMBL of both oceans, they were detected in only one surface water sample, suggesting no preferential proliferation there (Fig. 4f). Nevertheless, it cannot be excluded that the atmospheric microbial community could potentially affect bacterial composition and function in the surface water upon deposition.
In addition, we suggest that the local marine bacterial community emitted into the AMBL is enriched by long-range transported bacteria, associated with particle transport from terrestrial ecosystems (Fig. 4g). This observation is more evident in the Atlantic AMBL, where the microbial composition of Atlantic aerosols is associated with terrestrial and in particular, dust sources (e.g., Methylobacterium, Mesorhizobium; Fig. 4h).
This study depicts a high-resolution spatial diversity of airborne bacteria and their shifts between the ocean and the overlying atmosphere during the northern hemisphere springtime at both local and global scales. The interplay between the ocean surface and atmospheric feedback provides new opportunities for future studies to further explore the selective properties of marine microbes within both environments and how they in turn may affect biogeochemical cycles.
Methods
Sampling
Details of the expedition and sampling system used are described extensively in Flores et al.50. In short, marine aerosols were collected aboard the R/V Tara during the first year of the Tara Pacific Expedition58. Airborne particles were collected at ~15 m above sea level during the Atlantic crossing from Lorient, France, to Miami, USA. After Miami, the inlet was relocated to ~30 m above sea level. The inlet was constructed out of conductive tubing of 1.9 cm inner diameter and a funnel (allowing the collection of all diameters) and mounted on the rear backstay of Tara. A custom-made aerosol filter system consisting of four 47 mm filter holders and one vacuum pump (Diaphragm pump ME 16 NT, Vacuubrand BmbH & Co KG, Wertheim, Germany) was installed at the end of the inlet and used to collect the marine particles.
The flow through the filter system was 80 lpm (20 lpm for each filter) during the Atlantic crossing and 120 lpm (30 lpm for each filter) for the Pacific crossing. Three of the four filter holders were loaded with PVDF filters (47 mm diameter, 0.45 μm pore size, PAL, Port Washington, New York, USA), and were used for the current study. The flow rates of each filter holder were monitored continuously and recorded at the beginning and the end of each sampling event. Aerosols were collected for periods between approximately 12–24 h (see Supplementary Data 1 for the exact times). The filters were folded into a 2 ml cryotube and immediately dropped into liquid nitrogen, and these conditions were maintained while on board. Blank filters were collected by placing filters on the filter holders, closing them, reopening the holders, folding the filters into cryotubes, and dropping them into liquid nitrogen. These conditions were maintained while on board. Thorough validation tests were conducted to verify no boat-originated contamination masked the airborne bacterial population composition (See Supplementary Note 2, 6). The surface water sample collection is described in detail in Gorsky et al.58. In short, a “Dolphin sampler”, collecting <2000 μm size particles of the ocean surface water, and connected to a large volume peristaltic pump installed on the deck (max flow rate = 3 m3 h–1), was used for water sampling. Each sample endured for ~120 min. Water serial filtration (<0.22, 0.22–3, and 3–20 µm) was performed using 142 mm diameter polycarbonate filters (Millipore, Burlington, Massachusetts, USA), and the 0.22–3 µm fraction, where free bacteria are expected to be concentrated, was used for the current study. The filters were folded into a 5 ml cryotube and immediately dropped into liquid nitrogen, and these conditions were maintained while on board. The samples were shipped to the laboratory on dry ice and kept at −80 °C until DNA extraction was carried in the laboratory.
Back-trajectory analysis
The air mass origins were tracked using the National Oceanic and Atmospheric Administration HYSPLIT trajectory model and Global Data Assimilation System meteorological data59. Although the average residence time for 3 µm particles is of about 4.7 days5, as a considerable terrestrial influence was observed, the model was run to obtain the 240-h (10 days) back trajectories using the “Ensemble option” at an endpoint height of 250 m, which is the minimum height for an optimal configuration of the ensemble. The presented back trajectories in Fig. 1 are the average of the 27 back trajectories produced by the Ensemble option.
DNA extraction and sequencing
DNA extraction from 0.22–3 μm water filters was performed as described in Alberti et al.60. Extraction of DNA from the air and Blank filters, with minute amounts of DNA, was carried out using the DNeasy PowerWater Kit (Qiagen, Hilden, Germany), following a thorough optimization procedure for the extraction of low DNA concentrations from air filters (see Supplementary Note 1 and Fig. S7). To avoid a covariant effect on the results, we extracted DNA from both air and blank samples without knowing their collection date, location, or type (i.e., air or blank sample). Each sample was labeled with a numbered barcode, and the date and sampling data were specified only in the metadata. This allowed randomizing sample order in extraction and analysis. The processing of the water and air samples was performed separately, with random DNA extraction order for the different transects, thus preventing cross-contaminations between the different environments and geographic locations.
The DNA concentrations were evaluated with a Qubit® 3.0 Fluorometer (Thermo Fischer, Massachusetts, USA), using the DeNovix (Wilmington, Delaware, USA) dsDNA Ultra High Sensitivity Assay. For DNA sequencing, the bacterial V4–V5 region of the 16S rRNA gene (515Fa: 5′–GTGYCAGCMGCCGCGGTAA–3′, and 926 R: 5′–CCGYCAATTYMTTTRAGTTT–3′)61, was amplified. A PCR mix of 25 μl was prepared in triplicate using 1X Mytaq mix (Bioline, London, UK), 0.2 μM primers, 4 μl DNA extract, and PCR-grade water (Merck, Darmstadt, Germany). A no-template control was included in all runs, as well as DNA from a mock community (ZymoBIOMICS Microbial Community DNA Standard; Zymo, Irvine, CA, USA). The PCR products were validated on 2% agarose gel, and triplicates were pooled and sent to Genoscope, the French National DNA Sequencing Facility. The PCR products of the water samples from the Pacific transect were sent to the DNA Sequencing Facility at the University of Illinois at Chicago.
DNA sequencing in both facilities was conducted using Illumina MiSeq sequencing technology (maximum read length of 2 × 300 base pairs). No batch effects were observed, as validated by mock community positive control sequencing.
Sequences processing
Raw amplicon reads provided by the Genoscope sequencing facility were first merged using usearch (v11, -fastq_mergepairs; -fastq_pctid 90 commands)62, followed by the DADA2 pipeline (V 1.12)63, using R (dada2 package). The merged reads (provided as.fastq files) were trimmed and filtered by removing reads exceeding the maximum expected error of 2 bp or an ambiguous read. The reads were dereplicated to acquire unique sequences, which were used to infer sequence variants with the trained error model. After chimeric sequence removal, ASVs were used to assign taxa. ASVs were taxonomically assigned using dada2, with two steps: classification of sequences against the SILVA training dataset (nr_V 132, https://doi.org/10.5281/zenodo.1172783), followed by an exact matching between ASVs and the SILVA species assignment dataset (V 132) providing species-level assignment64. Raw reads provided by the sequencing facility of the University of Illinois were also processed using the dada2 pipeline. Forward and reversed reads were merged after trimming and filtering steps. Dereplication and annotation processes were carried out as described above. A total of 11,577 bacterial ASVs were obtained after error correction, chimera, Archean, chloroplast, and mitochondrial DNA removal. Possible shifts in microbial composition due to sequence facility variation were excluded by comparing mock community positive controls sequenced and analyzed in both sequencing facilities and pipelines described above. The microbial population sequencing analysis was performed only after a thorough decontamination procedure, to mitigate putative contamination in sequence libraries from the air samples with low microbial biomass65, as described in Supplementary Note 3.
Calculating autotrophs/heterotrophs rates
The ratio of known autotrophic to heterotrophic bacterial ASV was calculated, to estimate the fractal primary production potential in the AMBL25. A list of the autotrophic bacteria we referred to in our analysis is detailed in the SI (Table S5).
Environmental ontology
Environmental descriptive terms were extracted from the closest matches (97% identity) using the SEQenv pipeline for Python (version 1.3.0) with default parameters66 and ENVO terms. The input data included FASTA files of sequences after quality control check and removal of blank contaminants per each sample, to be compared to highly similar sequences from public repositories (such as GenBank, using the NCBI nucleotide database). The unassigned ASVs were removed from the analysis and included 26.4 and 27.6% for the Atlantic and Pacific air, respectively, and 10 and 28% for the Atlantic and Pacific water, respectively. The Firmicutes assignment of ENVO terms was the lowest, with 52 and 57% non-assigned ASVs in the Atlantic and Pacific Firmicutes, respectively. The relative abundance of the ASVs associated with several ENVO terms were multiplied by the relative contribution of each term, as specified in Table S7, S8 (https://doi.org/10.6084/m9.figshare.19354610.v1). The terms were further clustered into five main groups: marine, terrestrial, freshwater, anthropogenic, and unclassified, as detailed in Supplementary Data 6. A detailed description of the pipeline flow is given in Supplementary Note 7.
Statistical analysis
Statistical and communal-diversity analyses were conducted using R software. The ASV analysis was performed using phyloseq (V. 1.36.0)67. Analysis of similarities (ANOSIM, vegan V. 2.5-7)68 was used to verify the significance of the nonmetric multidimensional scaling (NMDS) ordination for taxonomic grouping (using the Bray-Curtis dissimilarity score), and differences in phyla composition between environments. The CLR-transformation of ASV abundance into differential abundance was conducted (compositions, V. 2.0-2), following zero-counts adjusted (zCompositions, V. 1.3.4). Analysis of molecular variance (AMOVA, ade4 V. 1.7-17)69 was used to verify the significance of the Bray-Curtis dissimilarity differences in bacterial composition between biomes. Alpha diversity values were represented by the Shannon index and were tested for differences using the Wilcoxon rank test (DivNet V. 0.4.0)70. Most likelihood phylogenetic trees were constructed based on bacterial ASVs observed in more than one sample. The trees were drawn using the interactive tree of life tool, iTOL, from a.tre file prepared from aligned sequences in R using the ape (V. 5.5)71 package (with the Neighbor-Joining Tree Estimation function; njs). Significance in the association between bacterial taxa and environmental variables was tested using MaAsLin2 (V. 1.6.0)72.
Data availability
The raw 16S amplicon sequences were deposited in the European Nucleotide Archive at EMBL-EBI (accession numbers: PRJEB39048 and PRJEB38899). Tables S7 and S8 were deposited in Figshare.com (https://doi.org/10.6084/m9.figshare.19354610.v1).
References
Bar-On, Y. M., Phillips, R. & Milo, R. The biomass distribution on Earth. Proc Natl Acad Sci USA 115, 6506–6511 (2018).
Sunagawa, S. et al. Structure and function of the global ocean microbiome. Science 348, 1261359 (2015).
Fröhlich-Nowoisky, J. et al. Bioaerosols in the Earth system: climate, health, and ecosystem interactions. Atmos. Res. 182, 346–376 (2016).
Ruiz-Gil, T. et al. Airborne bacterial communities of outdoor environments and their associated influencing factors. Environ. Int. 145, 106156 (2020).
Burrows, S. M., Elbert, W., Lawrence, M. G. & Pöschl, U. Bacteria in the global atmosphere—Part 1: review and synthesis of literature data for different ecosystems. Atmos. Chem. Phys. 9, 9263–9280 (2009).
Alsante, A. N., Thornton, D. C. O. & Brooks, S. D. Ocean aerobiology. Front. Microbiol. https://doi.org/10.3389/fmicb.2021.764178 (2021).
Baas Becking, L. G. M. Geobiologie of Inleiding tot de Milieukunde (W.P. Van Stockum & Zoon, 1934).
Martiny, J. B. H. et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112 (2006).
Martin, K. et al. The biogeographic differentiation of algal microbiomes in the upper ocean from pole to pole. Nat. Commun. 12, 5483 (2021).
Zhou, J. & Ning, D. Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. https://doi.org/10.1128/mmbr.00002-17 (2017).
Rahav, E. et al. Airborne microbes contribute to N2 fixation in surface water of the Northern Red Sea. Geophys. Res. Lett. 45, 6186–6194 (2018).
Tang, W. et al. Widespread phytoplankton blooms triggered by 2019–2020 Australian wildfires. Nature 597, 370–375 (2021).
DeLong, E. F. et al. Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311, 496 (2006).
Gat, D., Mazar, Y., Cytryn, E. & Rudich, Y. Origin-dependent variations in the atmospheric microbiome community in Eastern Mediterranean dust storms. Environ. Sci. Technol. 51, 6709–6718 (2017).
Rahav, E., Belkin, N., Paytan, A. & Herut, B. The relationship between air-mass trajectories and the abundance of dust-borne prokaryotes at the SE Mediterranean Sea. Atmosphere 10, 280 (2019).
Maki, T. et al. Aeolian dispersal of bacteria associated with desert dust and anthropogenic particles over continental and oceanic surfaces. J. Geophys. Res. Atmos. 124, 5579–5588 (2019).
Archer, S. D. J. et al. Air mass source determines airborne microbial diversity at the ocean–atmosphere interface of the Great Barrier Reef marine ecosystem. ISME J. 14, 871–876 (2019).
Lang-Yona, N. et al. Links between airborne microbiome, meteorology, and chemical composition in northwestern Turkey. Sci. Total Environ. 725, 138227 (2020).
Amato, P. et al. Metatranscriptomic exploration of microbial functioning in clouds. Sci. Rep. 9, 4383 (2019).
Rodrigues, T. B. & Silva, A. E. T. Molecular Diversity of Environmental Prokaryotes (CRC Press, 2016).
Sul, W. J., Oliver, T. A., Ducklow, H. W., Amaral-Zettler, L. A. & Sogin, M. L. Marine bacteria exhibit a bipolar distribution. Proc. Natl Acad. Sci. USA 110, 2342–2347 (2013).
Quaiser, A. et al. Acidobacteria form a coherent but highly diverse group within the bacterial domain: evidence from environmental genomics. Mol. Microbiol. 50, 563–575 (2003).
Archer, S. D. J. et al. Airborne microbial transport limitation to isolated Antarctic soil habitats. Nat. Microbiol. 4, 925–932 (2019).
Caliz, J., Triado-Margarit, X., Camarero, L. & Casamayor, E. O. A long-term survey unveils strong seasonal patterns in the airborne microbiome coupled to general and regional atmospheric circulations. Proc. Natl Acad. Sci. USA 115, 12229–12234 (2018).
Lees, H. Biochemistry of Autotrophic Bacteria (Hassell Street Press, 2021).
Flombaum, P. et al. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl Acad. Sci. USA 110, 9824–9829 (2013).
DeVries, T. & Primeau, F. Dynamically and observationally constrained estimates of water-mass distributions and ages in the Global Ocean. J. Phys. Oceanogr. 41, 2381–2401 (2011).
Seinfeld, J. H. & Pandis, S. N. Atmospheric Chemistry and Physics: From Air Pollution to Climate Change (Wiley, 2016).
Huang, S. et al. Overview of biological ice nucleating particles in the atmosphere. Environ. Int. 146, 106197 (2021).
Siller, H., Rainey, F. A., Stackebrandt, E. & Winter, J. Isolation and characterization of a new gram-negative, acetone-degrading, nitrate-reducing bacterium from soil, Paracoccus solventivorans sp. nov. Int. J. Syst. Bacteriol. 46, 1125–1130 (1996).
Lee, J. H., Kim, Y. S., Choi, T. J., Lee, W. J. & Kim, Y. T. Paracoccus haeundaensis sp. nov., a Gram-negative, halophilic, astaxanthin-producing bacterium. Int. J. Syst. Evol. Microbiol. 54, 1699–1702 (2004).
Liu, X. Y., Wang, B. J., Jiang, C. Y. & Liu, S. J. Paracoccus sulfuroxidans sp. nov., a sulfur oxidizer from activated sludge. Int. J. Syst. Evol. Microbiol. 56, 2693–2695 (2006).
DeLeon-Rodriguez, N. et al. Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc. Natl Acad. Sci. USA 110, 2575–2580 (2013).
Mazar, Y., Cytryn, E., Erel, Y. & Rudich, Y. Effect of dust storms on the atmospheric microbiome in the Eastern Mediterranean. Environ. Sci. Technol. 50, 4194–4202 (2016).
Petroselli, C. et al. Characterization of long-range transported bioaerosols in the Central Mediterranean. Sci. Total Environ. 763, 143010–143010 (2021).
Ivanova, E. P. et al. Characterization of Bacillus strains of marine origin. Int. Microbiol. 2, 267–271 (1999).
Mondol, M. A. M., Shin, H. J. & Islam, M. T. Diversity of secondary metabolites from marine Bacillus species: chemistry and biological activity. Mar. Drugs 11, 2846–2872 (2013).
Fierer, N., Bradford, M. A. & Jackson, R. B. Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364 (2007).
Kim, J.-H. & Kim, W. Tumebacillus soli sp. nov., isolated from non-rhizosphere soil. Int. J. Syst. Evol. 66, 2192–2197 (2016).
Buttigieg, P. L. et al. The environment ontology: contextualising biological and biomedical entities. J. Biomed. Semant. 4, 43 (2013).
Mayol, E. et al. Long-range transport of airborne microbes over the global tropical and subtropical ocean. Nat. Commun. 8, 201 (2017).
Pootakham, W. et al. High resolution profiling of coral-associated bacterial communities using full-length 16S rRNA sequence data from PacBio SMRT sequencing system. Sci. Rep. 7, 2774 (2017).
Mescioglu, E. et al. Aerosol microbiome over the Mediterranean Sea diversity and abundance. Atmosphere 10, 440 (2019).
Chisholm, S. W. et al. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 334, 340–343 (1988).
Chisholm, S. W. et al. Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch. Microbiol. 157, 297–300 (1992).
Carareto Alves, L. M., de Souza, J. A. M., Varani, A. M. & Lemos, E. G. M. The Family Rhizobiaceae. In: The Prokaryotes. (eds Rosenberg, E., DeLong, E. F., Lory, S., Stackebrandt, E. & Thompson, F.) (Springer, 2014). https://doi.org/10.1007/978-3-642-30197-1_297.
Pujalte, M. J., Lucena, T., Ruvira, M. A., Arahal, D. R. & Macián, M. C. The Family Rhodobacteraceae. In: The Prokaryotes (eds Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E. & Thompson, F.) (Springer, 2014). https://doi.org/10.1007/978-3-642-30197-1_377.
Kelly, D. P., McDonald, I. R. & Wood, A. P. The Family Methylobacteriaceae. In The Prokaryotes (eds Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt E., & Thompson, F.) (Springer, 2014). https://doi.org/10.1007/978-3-642-30197-1_256.
González-Toril, E. et al. Impacts of Saharan dust intrusions on bacterial communities of the low troposphere. Sci. Rep. 10, 6837 (2020).
Flores, J. M. et al. Tara Pacific expedition’s atmospheric measurements. Marine aerosols across the Atlantic and Pacific Oceans overview and preliminary results. Bull. Am. Meteorol. Soc. 101, 536–554 (2020).
Flores, J. M. et al. Diel cycle of sea spray aerosol concentration. Nat. Commun. 12, 5476 (2021).
Kaufman, Y. J. et al. Dust transport and deposition observed from the Terra-Moderate Resolution Imaging Spectroradiometer (MODIS) spacecraft over the Atlantic Ocean. J. Geophys. Res. Atmos. https://doi.org/10.1029/2003jd004436 (2005).
Gama, C. et al. Seasonal patterns of Saharan dust over Cape Verde—a combined approach using observations and modelling. Tellus B Chem. Phys. Meteorol. 67, 24410 (2015).
Morris, R. M. et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810 (2002).
Fahlgren, C. et al. Seawater mesocosm experiments in the Arctic uncover differential transfer of marine bacteria to aerosols. Environ. Microbiol. Rep. 7, 460–470 (2015).
Wurl, O., Ekau, W., Landing, W. & Zappa, C. Sea surface microlayer in a changing ocean—A perspective. Elementa. Sci. Anthrop. https://doi.org/10.1525/elementa.228 (2017).
Michaud, J. M. et al. Taxon-specific aerosolization of bacteria and viruses in an experimental ocean-atmosphere mesocosm. Nat. Commun. 9, 2017 (2018).
Gorsky, G. et al. Expanding Tara Oceans protocols for underway, ecosystemic sampling of the ocean-atmosphere interface during Tara Pacific expedition (2016–2018). Front. Mar. Sci. 6, https://doi.org/10.3389/fmars.2019.00750 (2019).
Stein, A. F. et al. NOAA’s HYSPLIT atmospheric transport and dispersion modeling system. Bull. Am. Meteorol. Soc. 96, 2059–2077 (2016).
Alberti, A. et al. Viral to metazoan marine plankton nucleotide sequences from the Tara Oceans expedition. Sci. Data 4, 170093 (2017).
Parada, A. E., Needham, D. M. & Fuhrman, J. A. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414 (2016).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Eisenhofer, R. et al. Contamination in low microbial biomass microbiome studies: issues and recommendations. Trends Microbiol. 27, 105–117 (2019).
Ijaz, A. Z., Jeffries, T. C., Ijaz, U. Z., Hamonts, K. & Singh, B. K. Extending SEQenv: a taxa-centric approach to environmental annotations of 16S rDNA sequences. PeerJ 5, e3827–e3827 (2017).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE https://doi.org/10.1371/journal.pone.0061217 (2013).
Oksanen, J. et al. Vegan: community ecology package (version 2.5-6). The Comprehensive R Archive Network. 2019.
Dray, S. & Dufour, A.-B. The ade4 package: implementing the duality diagram for ecologists. J. Stat. Softw. 22, 1–20 (2007).
Willis, A. D. & Martin, B. D. Estimating diversity in networked ecological communities. Biostatistics 23, 207–222 (2022).
Paradis, E. & Schliep, K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2018).
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 17, e1009442 (2021).
Acknowledgements
This research was supported by a research grant from Scott Jordan and Gina Valdez, the De Botton for Marine Science, the Yeda-Sela Center for Basic Research, and the Sustainability and Energy Research Initiative (SAERI). We are grateful to the following institutions for their financial and scientific support in Tara Pacific expedition: CNRS, PSL, CSM, EPHE, Genoscope/CEA, and France Génomique funding (ANR-10-INBS-09-08), Inserm, Université Côte d’Azur, ANR, agnès b., UNESCO-IOC, the Veolia Environment Foundation, Région Bretagne, Serge Ferrari, Billerudkorsnas, Amerisource Bergen Company, Lorient Agglomeration, Oceans by Disney, the Prince Albert II de Monaco Foundation, L’Oréal, Biotherm, France Collectivités, Kankyo Station, Fonds Français pour l’Environnement Mondial (FFEM), Etienne BOURGOIS, and the Tara Foundation teams and crew. The authors particularly thank Dr. Serge Planes, Prof. Denis Allemand, and the Tara Pacific consortium for their management and coordination. The authors gratefully acknowledging Prof. Daniel Sher for providing Prochlorococcus cultures for qPCR quantification, and Dr. Flora Vincent for her helpful, critical feedback on the manuscript. N.L.-Y. acknowledges support from the Women Bridging position and the Sustainability and Energy Research Initiative (SAERI), Weizmann Institute of Science. Y.R. acknowledges support from the Israel Science Foundation (grant #236/16). S.S. acknowledges support from the ETH and Helmut Horten Foundation. This is publication number # 15 of the Tara Pacific Consortium.
Author information
Authors and Affiliations
Contributions
Conceptualization: N.L.-Y., J.M.F, R.H., I.K., and A.V.; Methodology: N.L.-Y., R.H., A.A., J.P., C.B., and H.-J.R.; Investigation: N.L.-Y., J.M.F, I.K., and A.V.; Data curation: N.L.-Y., J.M.F, R.H., A.A., D.G., J.P., C.B., and H.-J.R.; Writing—original draft: N.L.-Y.; Visualization: N.L.-Y., J.M.F., M.T., I.K., and A.V.; Writing—review and editing: J.M.F., M.T., D.G., S.S., Y.R., I.K., and A.V.; Resources: P.W., S.S., Y.R., I.K., and A.V.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Earth & Environment thanks Kevin Lee and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Clare Davis.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lang-Yona, N., Flores, J.M., Haviv, R. et al. Terrestrial and marine influence on atmospheric bacterial diversity over the north Atlantic and Pacific Oceans. Commun Earth Environ 3, 121 (2022). https://doi.org/10.1038/s43247-022-00441-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43247-022-00441-6
This article is cited by
-
Marine viruses disperse bidirectionally along the natural water cycle
Nature Communications (2023)
-
Ice nucleation catalyzed by the photosynthesis enzyme RuBisCO and other abundant biomolecules
Communications Earth & Environment (2023)
-
Regionally sourced bioaerosols drive high-temperature ice nucleating particles in the Arctic
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.